The hubbub about the heart’s lub-dub
After a decade of debate, Brandeis scientists have a clear picture of a controversial protein
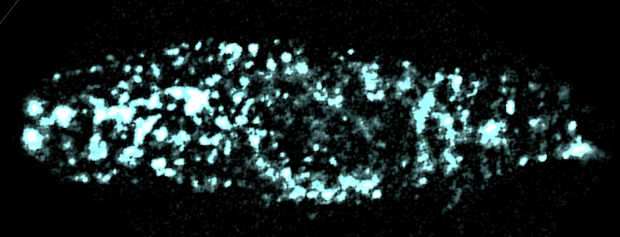
E1 protein, on the surface of a mammalian cell, illuminated with a teal fluorescent protein and viewed through a laser mounted total internal reflection microscope.
For years, scientists have debated how many KCNE1 proteins are required to build one of these channels, theorizing anywhere between one and 14. Now, Brandeis researchers found that these channels are built with two E1s. Understanding the construction of this channel is key to understanding life-threatening heart conditions, such as arrhythmias, and developing drugs to threat those conditions.
Leigh Plant, assistant research professor of biochemistry, along with postdoctoral fellows Dazhi Xiong, Hui Dai and provost and professor of biochemistry Steve Goldstein, published their findings in the Proceedings of the National Academy of Sciences on Monday, March 3.
This report challenges a previous study — the findings of which are currently being used in drug development trials and animal models — that anywhere between one and four E1s are required per channel. Brandeis researchers hope their new findings may help create more effective models to study heart conditions and their treatment.
A single heartbeat is the slow expanding and contracting of the heart muscle. It is controlled, in part, by a series of channels on the surface of heart cells that regulate the movement of different ions into and out of the cells. The potassium ion channel is critical to ending each heart contraction and is made up of the proteins Q1 and E1. Q1s create the pore that the potassium flows through and the E1s control how slowly that pore opens and closes, how many channels are on the cell surface of each cell and how they are regulated by drugs.
Goldstein’s team observed E1 in live, mammalian cells at remarkable sensitivity, counting the proteins in individual channels, something that had never been done before in this area of research. Because this mechanism has been so widely debated, Goldstein and his team used three different means to count E1 — including tagging them with different fluorescent colors and using a scorpion toxin to bind to Q1. Each time, the team got the same results.
While there is always room for debate in science, Goldstein and his team said they hope these findings will give researchers a quintessential key to unlocking the intricacies of the heartbeat.
This research was funded by a grant from the National Institutes of Health.
Categories: Research, Science and Technology





