Research finds viruses can be defeated with their own engineering principles
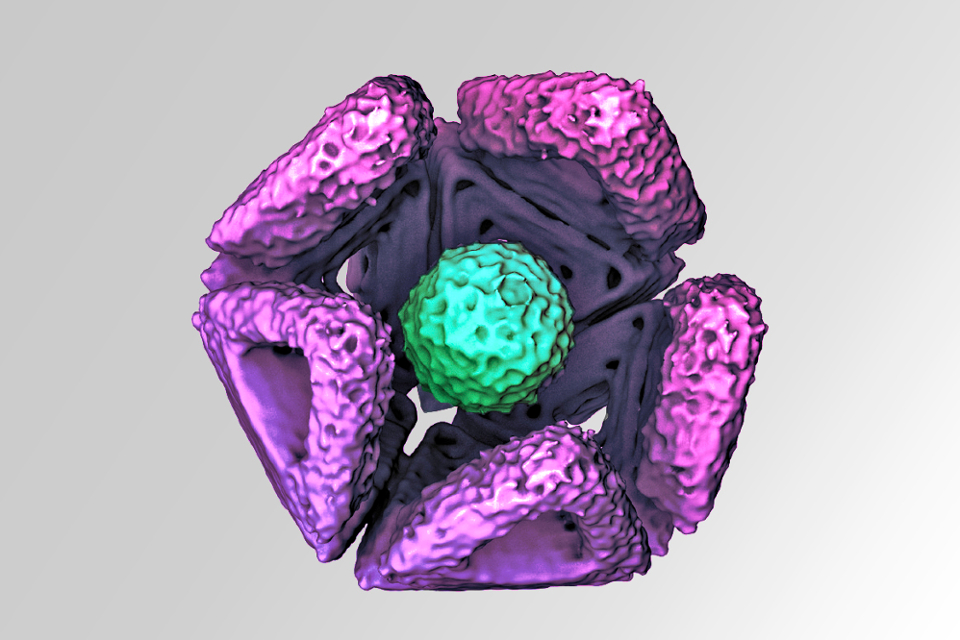
Caption: This cryo-electron microphotograph shows a pre-assembled icosahedral shell fragment of DNA that has trapped a hepatitis B virus core particle of 30 nm diameter. The DNA shell prevents the virus from fusing with the cell membrane, thereby preventing infection. https://doi.org/10.1038/s41563-021-01020-4. Color added for artistic effect, true color is unknown.
Brandeis researchers knew they had a major scientific accomplishment on their hands recently when, for the first time, they and their collaborators at the Technical University of Munich were able to program bits of DNA strands to fold up and self-assemble into predetermined complex geometric shapes.
The tragedy of the global COVID-19 pandemic, however, presented the team with a moonshot idea: using those tiny geometric structures to act as an antiviral agent. And it worked.
“It’s beyond my wildest dreams that this would work,” said Seth Fraden, director of Brandeis’ Materials Research Science and Engineering Center. “It’s just so exciting...I can’t sleep at night thinking about what we can do with this stuff.”
Fraden, along with Michael Hagan, a physics and quantitative biology professor at Brandeis, detailed the breakthrough in a new paper published in Nature Materials with their research partners at the Technical University of Munich.
The trans-Atlantic team used microscopic, triangular building blocks to create shells capable of capturing viruses and neutralizing them. While the breakthrough isn’t nearly ready for patient trials yet, it opens a potential new avenue for treating the deadly pathogens, administering drugs, or delivering gene therapies.
“This is a positive test of the theory and takes a big step towards eventually having those applications,” said Hagan.
In a basic sense, the achievement amounts to humans figuring out how to do something viruses — which are some of earth’s most primitive biological structures — have been doing for a billion years, Fraden said. Scientists have known for decades that viruses are able to contort and assemble themselves into highly-efficient structures that can wreak havoc on host bodies. Figuring out what makes this behavior possible, from an engineering standpoint, is something the Brandeis team has been working on for years.
“We asked, ‘OK, if viruses use these very efficient engineering principles to assemble themselves, why can’t we as material scientists make nanostructures based on the same engineering principles that the viruses use?’” Fraden said.
Hagan said that based on models and simulations they had developed, they felt they have made significant advances toward figuring out this complex biological puzzle - at least theoretically.
“You don’t really understand something unless you’re able to take those design principles and from scratch build a system based on those principles and see if it works,” he said.
Fraden’s team, which had previously partnered with Hendrik Dietz at the Technical University of Munich on other nanotechnology projects, proposed exploring the creation of a programmable artificial shell platform in 2016.
The Brandeis team had previously come up with the theoretical idea of using triangles made up of DNA to efficiently construct various geometric shapes — think Buckminster Fuller’s famous geodesic dome. The trick was demonstrating it is actually possible to program DNA to fold in specific ways, with specific binding structures in specific ranges, allowing the folded DNA to find each other and latch together like a self-assembling Lego structure.
The work is built on principles elucidated by Donald Caspar, a Brandeis emeritus professor in biology, who coined the term “structural biology” and established a lab at Brandeis in the 1970s that studied the molecular structure of viruses.
Fraden said Dietz’s team at the Technology University of Munich are world leaders in the field of nanobiotechnology known as “DNA origami," or how to design DNA to fold in complex 3D forms.
“Combining their expertise in origami with our expertise in self-assembly led to incredibly rapid advances,” Hagen said.
Then COVID-19 struck, and the Munich researchers realized the potential value of expanding the experiment to see if shells could be constructed capable of effectively placing viruses in quarantine. Researchers programmed the DNA to create icosahedron-shaped capsids with apertures just wide enough to engulf a virus - in this case a hepatitis B or adeno-associated virus.
Much like an insect entering a pitcher plant, Fraden said, once inside the shell, the virus can’t escape. Antibodies applied to the shell’s interior kept the viruses bound inside; the shell also acted as a protective bumper, preventing the virus’ spike proteins from latching onto a host.
“We achieved near complete inactivation by engulfing [the hepatitis B virus] in a surrounding shell in vitro and could also effectively block [the adeno-associated virus] from infecting live cells,” the team wrote in its paper.
“It worked especially well, way beyond what we imagined possible,” Fraden said.
While the innovation has multiple possible implications for nanotechnologies and material sciences, it’s the biopharma side that really has researchers jazzed. For example, antibiotics effectively kill bacterial infections, even while the infection is active in a patient. Vaccinations trigger an immune system response in a patient before they get contract a virus, and some antivirals interrupt replication during active infections. But there currently is no effective treatment that reduces viral load. A high viral load can increase transmission risk and the severity of the illness with some viruses, such as COVID-19.
“If you have a heavy load of virus in your body, this is a potential platform to effectively sweep your body clean of the virus,” Fraden said. “There aren’t any generic methods that do that.”
Hagen said the immediate next steps involve using this newly proven testing process to fine tune models and improve design efficiency.
“It’s super exciting,” he said.
Categories: Research, Science and Technology





