Jeff Gelles

Aron and Imre Tauber Professor of Biochemistry and Molecular Pharmacology
Research Description
Single-molecule Biochemistry and Biophysics. Transcription and RNA processing. Cytoskeletal networks and regulation.
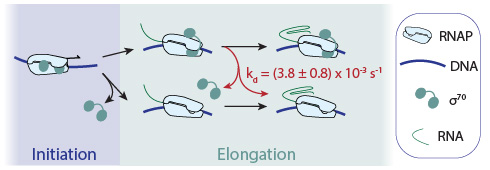
Living cells are chock full of dynamic complexes of protein, RNA, and DNA molecules. We want to understand the fundamental chemical and physical mechanisms through which these molecular machines perform essential biological processes. Our research is focused in two different areas of study: 1) the function of the molecular machines essential to gene expression and its regulation, in particular those that control the synthesis and processing of messenger RNAs, and 2) the function of molecular machines essential to the organization and utilization of the actin and microtubule cytoskeleton of eukaryotic cells.
All of the processes that we study involve dynamic molecular assemblies and multiple reaction intermediates. It is challenging to study their mechanisms using conventional biochemical approaches because different individual molecules in a population are doing different things at the same time. To overcome this “crowd noise problem”, we have developed and used single-molecule light microscopy methods that allow us to observe the behavior of isolated individual molecules and molecular complexes in real time. Using these techniques we can directly observe the assembly, rearrangements, and disassembly of molecular complexes, characterize conformational changes, and detect biochemical reactions. Together, these methods permit comprehensive investigation of molecular mechanisms, revealing reaction pathways and allowing quantitative evaluation and modeling of reaction dynamics. We perform these single-molecule experiments both in systems reconstituted from purified componentsand in extracts that recapitulate the molecular complexity of the living cell.

Our research uses approaches from scientific fields ranging from cell biology and genetics through biochemistry and molecular biology to physical chemistry and condensed-matter physics. In our experience, the most exciting advances in science often arise when scientists from different disciplines collaborate.