Dorothee Kern
 Professor Emeritus of Biochemistry
Professor Emeritus of Biochemistry
Howard Hughes Medical Institute Investigator
Research Description
Dynamics of enzymes. Magnetic resonance methods.
To get a personal flair of our current research, check out these two iBiology videos. iBiology’s mission is to bring the worlds best biology to you.
My lab uses biophysical analytical techniques to unravel the dynamic personality of enzymes, signaling proteins, and the molecules they affect. We are particularly interested in the evolution of the impressive catalytic power of enzymes and the evolution of more complex signaling features in higher organisms. To shed light on these fundamental questions, modern and resurrected ancestral proteins are being studied with atomistic resolution by combining experiments and computation.
A key feature of life is change over time. In a search for how and why biological processes happen, my colleagues and I study, at the molecular level, changes of atomic coordinates in proteins over time. The ultimate goal is to "visualize" proteins at atomic resolution in real time as they function: enzymes during catalysis, signaling proteins in action, and proteins and drugs binding to their partners. To accomplish this, we are using a variety of biophysical methods, including NMR (nuclear magnetic resonance) spectroscopy, x-ray crystallography, fast kinetics, single-molecule FRET (fluorescence resonance energy transfer), molecular dynamics simulations, bioinformatics and other computational approaches. We then build the bridge from the microscopic dynamic behavior of individual proteins to the macroscopic dynamic behavior of biological function. Ultimately we employ the knowledge of protein dynamics for the design of novel highly efficient inhibitors.
The 2 substrate molecules in green are “hidden” in the active site in the closed state where phosphoryl-transfer happens. The visualized opening is the rate-limiting step for this kinase.
Enzymes in Action
That enzymatic activity requires a precise balance between flexibility and stability is a widely accepted concept. However, key questions remain: How do motions on different timescales relate to each other and contribute to this balance? Have proteins evolved so that substates necessary for activity are preferably accessible?
We have developed approaches that allow quantitative and residue-specific measurements of dynamics in enzymes during catalysis by NMR spectroscopy. Strikingly, we found for several enzymes that the rate-limiting step for the overall turnover is a conformational change and not the chemical step. In other words, nature has "perfectionized" the enzyme to lower the energy barrier for chemistry, but collective motions are the price to be paid.
Moreover, we recently showed that motions in enzymes are not random but preferentially follow the pathways that create the configuration capable of proficient chemistry. This situation is analogous to protein folding, which is biased to sample only a small portion of the energy landscape. The expansion of the concept of nonrandom sampling of conformational space for efficient biological function—from folding to conformational rearrangements within the folded space—combines both phenomena through the energy landscape.
We are also investigating the hierarchy in space and time for protein dynamics. By comparing a mesophilic and hyperthermophilic enzyme pair, we identified a linkage between three different "tiers" of dynamic timescales: (1) thermally driven, fast (picosecond), local atomic fluctuations; (2) faster (nanosecond) motions of whole segments; and (3) larger amplitude, collective, slower motions (microsecond to millisecond), the timescale of catalysis. Importantly, we were able to demonstrate that those dynamic features are directly connected to function.
We propose that the pre-sampling of conformational states needed for catalysis and selective binding of substrates to these substates might be a general paradigm of enzyme catalysis. We are attacking the next immediate questions: How does a protein move from one energy valley into another, and what are the pathways and the transition states? Can minor conformational substates be predicted from known structures? Can we apply the knowledge gained about physical principles of proteins to design proteins with desired functions? Can we successfully incorporate a dynamic view into rational drug design?
Evolution of Enzymatic Power and Complex Signaling
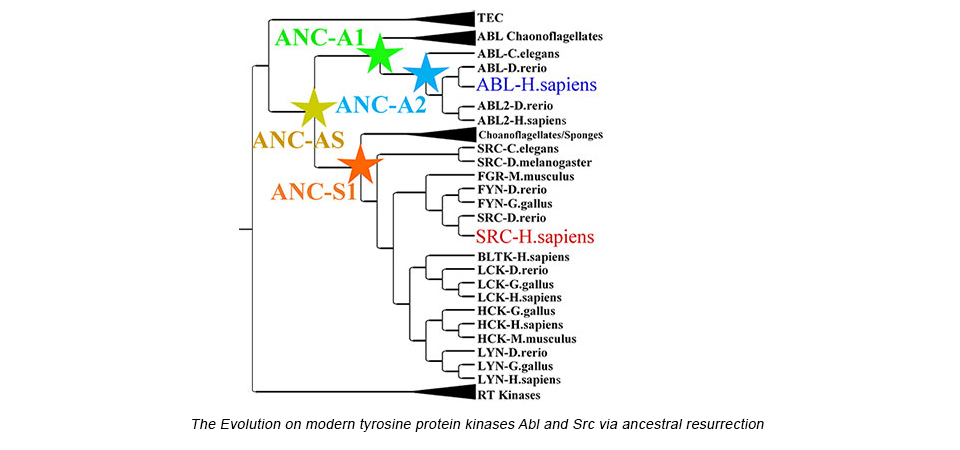
Our success in characterizing energy landscapes of enzymes and our first steps toward understanding the molecular pathways of conformational transitions have prompted us to begin applying these lessons to the development of artificial enzymes. While these first designer enzymes are a breakthrough, their catalytic turnover rates have been very low, most likely because we do not yet understand the defined and efficient changes in structure during catalysis. To address this principal question, we recapitulate a proven successful "design" of enzymes, the work of natural evolution. While the calculation of phylogenetic trees has become very popular (and meaningful) in the last years due to the huge explosion of sequences of diverse organisms, many basic questions of molecular evolution are unresolved, such as (1) Were the ancestors promiscuous enzymes from which specificity evolved? (2) Would it therefore be easier to design desired function from ancestors? Almost completely missing in the evolutionary field is the detailed biophysical characterization of ancestors and the lineage from those to our modern enzymes. Our first resurrected enzymes (up to ~3 billion years back) are not only active, but indeed give clues about the evolution of protein dynamics as an essential part of catalysis.
Molecular Mechanism of Phosphorylation-Mediated Signaling
Phosphorylation is a widely used mechanism in signaling. Through a combination of NMR structural and dynamic experiments, we found that signaling does not work through a mechanism like a light switch (on, off) but rather through a population-shift mechanism; the active state is already populated to a low percentage before phosphorylation, and phosphorylation shifts this preexisting equilibrium. In other words, this sampling of substates is essential for phosphorylation, shifting the textbook paradigm of phosphorylation inducing a new structure. We are exploring the energy landscape for this signaling protein, including both the inactive/active interconversion and the folding. One key question in biology is how proteins can quickly change among distinct structures (needed for function) without unfolding. We anticipate that lessons learned for this kinase are widely applicable to other kinases.
Mechanism of Signaling Mediated by Prolyl Isomerization
Prolyl isomerases, the enzymes that catalyze the reversible cis/trans isomerization of prolyl peptide bonds, have been implicated to play a crucial role in many biological processes, including cell cycle control, ion channel conductance, cell signaling, neurodegeneration, cancer, transcription, and HIV virulence. However, the molecular mechanisms by which these enzymes affect such a variety of biological processes are poorly understood. We have used NMR spectroscopy together with in vivo experiments to shed light into the molecular mechanism of how these enzymes, which do not break or make bonds but just accelerate a 180° rotation about the peptide bond, control cell cycle, cocaine addiction via metatropic glutamate receptor, chemotropic nerve guidance, and tubulin dynamics through the Alzheimer's protein tau. Catalysis of the natively folded protein substrates, such as the HIV capsid, the Trp channel, the synaptic receptor mGluR, and tau are indeed the trigger for the found phenotypes. In a search for new inhibitors, we are characterizing the energy landscapes for human CypA and human Pin1, two of the most important prolyl isomerases.
Selected Publications
- Hadzipasic A, Wilson C, Nguyen V, Kern N, Kim C, Pitsawong W, Villali J, Zheng Y & Kern D. Ancient origins of allosteric activation in a Ser-Thr kinase. Science. 2020 Feb 21;367(6480):912-917.
- Otten R, Pádua RAP, Bunzel HA, Nguyen V, Pitsawong W, Patterson M, Sui S, Perry SL, Cohen AE, Hilvert D & Kern D (2020). How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science. 2020 Dec 18;370(6523):1442-1446.
- Hoemberger M, Pitsawong W & Kern D. Cumulative mechanism of several major imatinib-resistant mutations in Abl kinase (2020). Proc Natl Acad Sci USA. 2020 Aug 11;117(32):19221-19227.
- Pádua AP, Sun Y, Marko I, Pitsawong W, Stiller JB, Otten R and Kern D. Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2. Nat Commun. 2018 Oct 30;9(1):4507. doi: 10.1038/s41467-018-06814-w
- Pitsawong W, Buosi V, Otten R, Agafonov RV, Zorba A, Kern N, Kutter S, Kern G, Pádua RAP, Meniche X and Kern D. Dynamics of human protein kinases Aurora A linked to drug selectivity. 2018, bioRxiv 286310; doi: https://doi.org/10.1101/286310, eLife. 2018; 7: e36656
- Otten R, Liu L, Kenner LR, Clarkson MW, Mavor D, Tawfik DS, Kern D and Fraser JS (2018). Rescue of Conformational Dynamics in Enzyme Catalysis by Directed Evolution. Nat Commun. 2018 Apr 3;9(1):1314. doi: 10.1038/s41467-018-03562-9.
- Nguyen V, Wilson C, Hoemberger M, Stiller JB, Agafonov RV, Kutter S, English J, Theobald DL, Kern D (2017). Evolutionary drivers of thermoadaptation in enzyme catalysis. Science. 2017 Jan 20;355(6322):289-294.
- Eichner T, Kutter S, Labeikovsky W, Buosi V, Kern D (2016). Molecular Mechanism of Pin1-Tau Recognition and Catalysis. J Mol Biol. 2016 May 8;428(9 Pt A):1760-75.
- Kutter S, Eichner T, Deaconescu AM, Kern D (2016). Regulation of Microtubule Assembly by Tau and not by Pin1. J Mol Biol. 2016 May 8;428(9 Pt A):1742-59.
- Wilson C, Agafonov RV, Kern D (2015). Drug targets evolve, and so should the methods. Mol Cell Oncol. 2015 Nov 11;3(4):e1046580.
- Chakrabarti KS, Agafonov RV, Pontiggia F, Otten R, Higgins MK, Schertler GF, Oprian DD, Kern D (2016). Conformational Selection in a Protein-Protein Interaction Revealed by Dynamic Pathway Analysis. Cell Reports. 2016 Jan 5;14(1):32-42.
- Pontiggia F, Pachov DV, Clarkson MW, Villali J, Hagan MF, Pande VS and Kern D (2015). Free energy landscape of activation in a signaling protein at atomic resolution. Nature Commun. 2015 June 15; 6:7284.
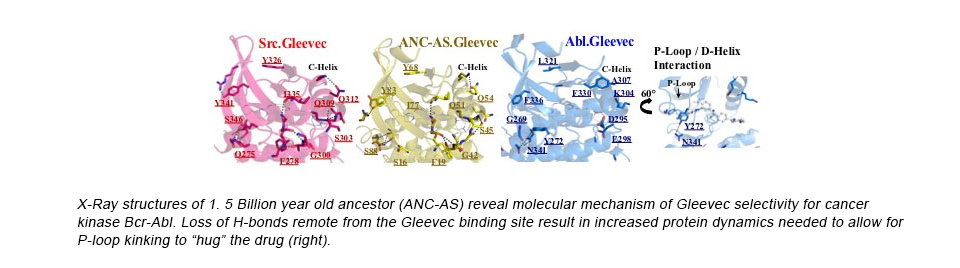
- Wilson C, Agafonov RV, Hoemberger M, Kutter S, Zorba A, Halpin J, Buosi V, Otten R, Waterman D, Theobald DL and Kern D (2015). Using Ancient Protein Kinases to Unravel a Modern Cancer Drug’s Mechanism. Science. Feb 20;347(6224):882-6.
-
Kerns SJ, Agafonov RV, Cho Y-J, Pontiggia F, Otten R, Pachov DV, Phung LA, Murphy PN, Thai V, Hagan MF, Alber T, Kern D. (2015) Energy landscape of Adenylate Kinase Catalysis. Nature Struct Mol Biol. 2015 22(2):124-31
- Agafonov RV, Wilson C, Otten R, Buosi V, Kern D (2014) .Energetic dissection of Gleevec’s selectivity towards human tyrosine kinases. Nature Struct Mol Biol. 2014 Oct;21(10):848-53.
- Zorba A, Buosi V, Kutter S, Kern N, Pontiggia F, Cho YJ and Kern D (2014). Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. Elife. 2014, 27;3:e02667.
- Park JM, Hu JH, Milshteyn A, Zhang P-W, Moore CG, Park S, Datko MC, Doming RD, Reye CM, Wang XJ, Etzkorn FA, Xiao B, Szumlinski KK, Kern D, Linden DJ, Worley PF (2013). A Prolyl-isomerase Mediates Dopamine-dependent Plasticity and Cocaine Motor Sensitization. Cell. 2013 154(3):637-50.
- Morrison EA, DeKoster GT, Dutta S, Vafabakhsh R, Clarkson MW, Bahl A, Kern D, Ha T, Henzler-Wildman KA (2011). Antiparallel EmrE exports drugs by exchanging between asymmetric structures. Nature. 2011 Dec 18; 481, 45-50.
- Gardino AK, Villali J, Kivenson A, Lei M, Liu CF, Steindel P, Eisenmesser EZ, Labeikovsky W, Wolf-Watz M, Clarkson MW and Kern D (2009). Transient non-native bonds promote activation of a signaling protein. Cell. (2009), 139(6):1109-1118.
- Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T (2009). Hidden alternative structures of proline isomerase essential for catalysis. Nature. (2009), 462, 669-673.
- Henzler-Wildman KA and Kern D. Dynamic personalities of proteins. Nature. (2007) 450, 964-972.
- Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, Hübner CG, Kern D. Intrinsic motions along an enzymatic reaction trajectory. Nature. (2007) 450, 838-844.
- Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. (2007), 450: 913-916.
- Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, Skalicky JJ, Kay LE and Kern D. Intrinsic dynamics of an enzyme underlies catalysis. Nature. (2005), 438, 117-121.
- Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr Opin Struct Biol. (2003) 13(6):748-57.
- Eisenmesser EZ, Bosco DA, Akke M, and Kern D. Enzyme Dynamics During Catalysis. Science. (2002) 295:1520-3. Prof. Joseph J. Falke has written a perspective on this article.
- Volkman BF, Lipson D, Wemmer DE, Kern D. Two-State Allosteric Behavior in a Single-Domain Signaling Protein. Science. (2001) 291, 2429-2433.
- Kern D, Volkman BF, Luginbühl P, Nohaile MJ, Kustu S, and Wemmer DE. Structure of a transiently phosphorylated "switch" in bacterial signal transduction. Nature. (1999) 402, 894-898.
- Kern D, Kern G, Neef H, Tittmann K, Killenberg-Jabs M, Wikner C, Schneider G, & Hübner G. How thiamin diphosphate is activated in enzymes. Science. (1997) 275, 67-70.