Maria-Eirini Pandelia
 Associate Professor of Biochemistry
Associate Professor of Biochemistry
Research Description
Mapping the functional repertoire of (bio)inorganic systems and protein metallocofactors; paradigms of (bio)catalysts relevant to the human health and environment.
My research interests lie in the strong interplay between (bio)chemistry/(bio)catalysis, spectroscopy and kinetics, with particular focus on the dissection of the mechanistic determinants underlying medically and environmentally relevant enzymatic processes. My research relies on the synteny of bioinformatic, spectroscopic, structural and kinetic methods to identify and establish the modus operandii of metal-containing enzymes and delineate their catalytic and functional repertoire.
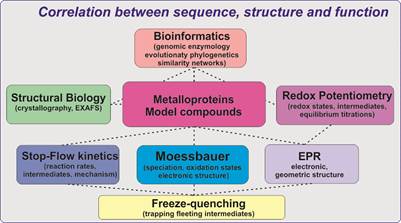
Establishing the operant mechanisms according to which (bio)catalysis takes place, consists of identifying the reactive species carrying out the ‘difficult’ chemistry. Because the enzymes that we are interested in not only play a key role in metabolic functions but also in industrially relevant applications, understanding their reactivity will allow us to direct and control these reactions. In most of the cases however the enzyme’s state that is responsible for catalysis is fleeting and cannot be characterized using conventional methods such as X-Ray crystallography. These short-lived intermediates can be identified and characterized by a combination of kinetic and spectroscopic approaches.
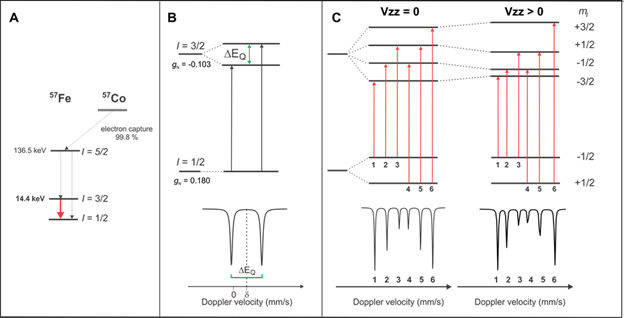
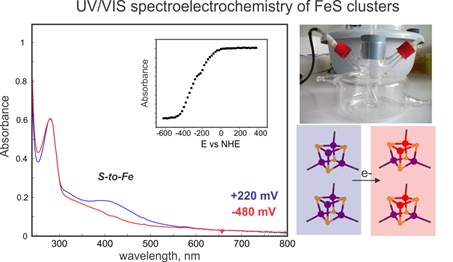
The main focuses of the research carried out comprise the use of Mössbauer and EPR spectroscopies coupled to time-resolved kinetics(optical) and redox potentiometry. In particular, 57Fe-Mössbauer plays a pivotal role among the many spectroscopic techniques that are routinely used to characterize Fe/S clusters and Fe-containing enzymes or bioinorganic synthetic compounds. Its strength lies in the fact that all chemically distinct 57Fe-labeled species in the sample are detected, irrespective of their oxidation and spin states. The Mössbauer spectrum is the weighted superposition of the individual subspectra of the various Fe sites contained in the sample and their relative areas are proportional to the relative concentration of the corresponding Fe species. For example, Fe/S clusters have unique Mössbauer-spectroscopic properties that allow for their identification and detailed characterization of their electronic and geometric structures.57Fe-Mössbauer spectroscopy when combined with Electron Paramagnetic Resonance (EPR) spectroscopy and analytical methods to determine the Fe:protein (synthetic compound) ratio represents the most rigorous method for establishing the types and stoichiometry of various Fe-containing cofactors.
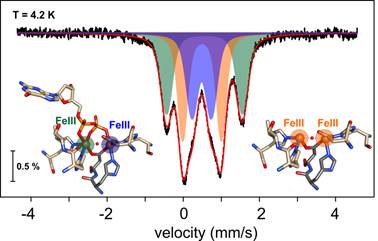
My work is centered on delineating the mechanisms according to which metalloproteins involved in processes essential for life perform the activation of small (or larger) molecules, how the specific identity of the metals in the active sites allows their chemical diversion and selectivity and what the functional role of iron-sulfur clusters in proteins involved in DNA synthesis and repair is. The research of the group revolves around proteins that are candidates to be the targets of new antiviral factors and to mediate DNA modifications and other important chemical transformations. These proteins often belong to large structural superfamilies with apparently similar representatives that are carrying out radically different reactions. We want to understand those functions and the extant determinants that endow this chemical versatility.
The experimental and theoretical approaches entail: transient and steady state kinetic properties (activity, rates, substrate binding and inhibition), thermodynamic properties (reduction potentials) under (non)catalytic conditions, electronic and chemical properties by Mössbauer/EPR spectroscopy, computational (bioinformatics) and structural biology (X-Ray crystallography, XAS methods) methods.
Selected Publications
- Characterization of Fe-S Clusters in Proteins by Mӧssbauer Spectroscopy. Ueda C, Langton M, Pandelia ME. Methods Mol Biol. 2021;2353:281-305. doi: 10.1007/978-1-0716-1605-5_15.
- The HD-Domain Metalloprotein Superfamily: An Apparent Common Protein Scaffold with Diverse Chemistries. Langton M, Sun S, Ueda C, Markey M, Chen J, Paddy I, Jiang P, Chin N, Milne A, Pandelia ME. Catalysts. 2020 Oct;10(10):1191. doi: 10.3390/catal10101191.
- Hepatitis B Virus Oncoprotein HBx Is Not an ATPase. Langton M, Pandelia ME. ACS Omega. 2020 Jun 28;5(27):16772-16778. eCollection 2020 Jul 14.
- HD-[HD-GYP] Phosphodiesterases: Activities and Evolutionary Diversification within the HD-GYP Family. Sun S, Pandelia ME. Biochemistry. 2020 Jun 30;59(25):2340-2350.
- Structures of Class Id Ribonucleotide Reductase Catalytic Subunits Reveal a Minimal Architecture for Deoxynucleotide Biosynthesis. Rose HR, Maggiolo AO, McBride MJ, Palowitch GM, Pandelia ME, Davis KM, Yennawar NH, Boal AK. Biochemistry. 2019 Apr 9;58(14):1845-1860.
- A New Microbial Pathway for Organophosphonate Degradation Catalyzed by Two Previously Misannotated Non-Heme-Iron Oxygenases. Rajakovich LJ, Pandelia ME, Mitchell AJ, Chang WC, Zhang B, Boal AK, Krebs C, Bollinger JM Jr. Biochemistry. 2019 Mar 26; 58(12):1627-1647.
- Spectroscopic and Electrochemical Characterization of the Mycofactocin Biosynthetic Protein, MftC, Provides Insight into Its Redox Flipping Mechanism. Ayikpoe R, Ngendahimana T, Langton M, Bonitatibus S, Walker LM, Eaton SS, Eaton GR, Pandelia ME, Elliott SJ, Latham JA. Biochemistry. 2019 Feb 19;58(7):940-950. doi: 10.1021/acs.biochem.8b01082.
- Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase. Wittenborn EC, Merrouch M, Ueda C, Fradale L, Léger C, Fourmond V, Pandelia ME, Dementin S, Drennan CL. Elife. 2018 Oct 2;7:e39451.
- Discovery and characterization of a prevalent human gut bacterial enzyme sufficient for the inactivation of a family of plant toxins. Koppel N, Bisanz JE, Pandelia ME, Turnbaugh PJ, Balskus EP. Elife. 2018 May 15;7:e33953. doi: 10.7554/eLife.33953.
- Structural Basis for Superoxide Activation of Flavobacterium johnsoniae Class I Ribonucleotide Reductase and for Radical Initiation by Its Dimanganese Cofactor. Rose HR, Ghosh MK, Maggiolo AO, Pollock CJ, Blaesi EJ, Hajj V, Wei Y, Rajakovich LJ, Chang WC, Han Y, Hajj M, Krebs C, Silakov A, Pandelia ME, Bollinger JM Jr, Boal AK. Biochemistry . 2018 May 8;57(18):2679-2693. doi: 10.1021/acs.biochem.8b00247.
- The biosynthesis of methanobactin. Kenney GE, Dassama LMK, Pandelia ME, Gizzi AS, Martinie RJ, Gao P, DeHart CJ, Schachner LF, Skinner OS, Ro SY, Zhu X, Sadek M, Thomas PM, Almo SC, Bollinger JM Jr, Krebs C, Kelleher NL, Rosenzweig AC. Science. 2018 Mar 23;359(6382):1411-1416.
- NADH reduction of nitroaromatics as a probe for residual ferric form high-spin in a cytochrome P450. Pochapsky TC, Wong N, Zhuang Y, Futcher J, Pandelia ME, Teitz DR, Colthart AM. Biochim Biophys Acta Proteins Proteom. 2018 Jan;1866(1):126-133.
-
Organometallic Complex Formed by an Unconventional Radical S-Adenosylmethionine Enzyme. Dong M, Horitani M, Dzikovski B, Pandelia ME, Krebs C, Freed JH, Hoffman BM, Lin H. J Am Chem Soc. 2016 Aug 10;138(31):9755-8.
-
Characterization of Lipoyl Synthase from Mycobacterium tuberculosis. Lanz ND, Lee KH, Horstmann AK, Pandelia ME, Cicchillo RM, Krebs C, Booker SJ (2016). Biochemistry. 2016 Mar 8;55(9):1372-83.
-
Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster. Aussignargues C, Pandelia ME, Sutter M, Plegaria JS, Zarzycki J, Turmo A, Huang J, Ducat DC, Hegg EL, Gibney BR, Kerfeld CA (2016). J Am Chem Soc. 2016 Apr 27;138(16):5262-70.
-
Pulse Double-Resonance EPR Techniques for the Study of Metallobiomolecules. Cox N, Nalepa A, Pandelia ME, Lubitz W, Savitsky A (2015). Methods Enzymol. 2015;563:211-49.
-
Rapid Reduction of the Diferric-Peroxyhemiacetal Intermediate in Aldehyde-Deformylating Oxygenase by a Cyanobacterial Ferredoxin: Evidence for a Free-Radical Mechanism. Rajakovich LJ, Nørgaard H, Warui DM, Chang WC, Li N, Booker SJ, Krebs C, Bollinger JM Jr, Pandelia ME (2015). J Am Chem Soc. 2015 Sep 16;137(36):11695-709.
-
The Carbon Monoxide Dehydrogenase from Desulfovibrio vulgaris. Hadj-Saïd J, Pandelia ME, Léger C, Fourmond V, Dementin S (2015). Biochim Biophys Acta. 2015 Dec;1847(12):1574-83.
-
Understanding the effect of monomeric iridium(III/IV) aquo complexes on the photoelectrochemistry of IrOx.nH2O-catalyzed water-splitting systems. Zhao Y, Vargas-Barbosa NM, Strayer ME, McCool NS, Pandelia ME, Saunders TP, Swierk JR, Callejas JF, Jensen L, Mallouk TE (2015). J Am Chem Soc. 2015 Jul 15;137(27):8749-57.
-
Mössbauer spectroscopy of Fe/S proteins. Pandelia ME, Lanz ND, Booker SJ, Krebs C (2015). Biochim Biophys Acta. 2015 Jun;1853(6):1395-1405.
-
Efficient delivery of long-chain fatty aldehydes from the Nostoc punctiforme acyl-acyl carrier protein reductase to its cognate aldehyde-deformylating oxygenase. Warui DM, Pandelia ME, Rajakovich LJ, Krebs C, Bollinger JM Jr, Booker SJ (2015). Biochemistry 2015 Feb 3;54(4):1006-15.
-
Evidence for a catalytically and kinetically competent enzyme-substrate cross-linked intermediate in catalysis by lipoyl synthase. Lanz ND, Pandelia ME, Kakar ES, Lee KH, Krebs C, Booker SJ (2014). Biochemistry 2014, 53(28):4557-72.
-
ChlR protein of Synechococcus sp. PCC 7002 is a transcription activator that uses an oxygen-sensitive [4Fe-4S] cluster to control genes involved in pigment biosynthesis. Ludwig M, Pandelia ME, Chew CY, Zhang B, Golbeck JH, Krebs C, Bryant DA (2014). J Biol Chem. 2014 Jun 13;289(24):16624-39.
-
Organophosphonate-degrading PhnZ reveals an emerging family of HD domain mixed-valent diiron oxygenases. Wörsdörfer B, Lingaraju M, Yennawar NH, Boal AK, Krebs C, Bollinger JM Jr, Pandelia ME (2013). Proc. Natl. Acad. Sci. USA, 2013, 110(47):18874-9.
-
Reply to Mouesca et al.: Electronic structure of the proximal [4Fe-3S] cluster of O2-tolerant [NiFe] hydrogenases. Pandelia ME, Bykov D, Izsak R, Infossi P, Giudici-Orticoni MT, Bill E, Neese F, Lubitz W (2013). Proc Natl Acad Sci USA. 2013 Jul 9;110(28):E2539.
-
Human anamorsin binds [2Fe-2S] clusters with unique electronic properties. Banci L, Ciofi-Baffoni S, Mikolajczyk M, Winkelmann J, Bill E, Pandelia ME (2013). J. Biol. Inorg. Chem., 2013, 18,pp 883-893.
-
A metal-metal bond in the light-induced state of [NiFe] hydrogenases with relevance to hydrogen evolution. Kampa M, Pandelia ME, Lubitz W, van Gastel M, Neese F (2013). J. Am. Chem. Soc., 2013, 135(10):3915-25.
-
Electronic structure of the unique [4Fe-3S] cluster in O2-tolerant hydrogenases characterized by 57Fe Mossbauer and EPR spectroscopy. Pandelia ME, Bykov D, Izsak R, Infossi P, Giudici-Orticoni MT, Bill E, Neese F, Lubitz W (2013). J. Am. Chem. Soc. 2013 135(42):15801-12.
-
Radical-translocation intermediates and hurdling of pathway defects in 'super-oxidized' (Mn(IV)/Fe(IV)) Chlamydia trachomatis ribonucleotide reductase. Dassama LM, Jiang W, Varano PT, Pandelia ME, Conner DA, Xie J, Bollinger JM Jr, Krebs C (2012). J. Am. Chem. Soc., 2012, 20498–20506.
-
Pulse Q-band EPR and ENDOR spectroscopies of the photochemically generated monoprotonated benzosemiquinone radical in frozen alcoholic solution. Flores M, Okamura MY, Niklas J, Pandelia ME, Lubitz W (2012). J Phys Chem B, 2012 August 2, 116(30):8890-900.
-
Evolution and diversification of Group 1 [NiFe] hydrogenases. Is there a phylogenetic marker for O(2)-tolerance? Pandelia ME, Lubitz W, Nitschke W (2012). Biochim Biophys Acta, 2012, 1817(9):1565-75.
-
Spectroscopic characterization of the key catalytic intermediate Ni-C in the O2-tolerant [NiFe] hydrogenase I from Aquifex aeolicus: evidence of a weakly bound hydride. Pandelia ME, Infossi P, Stein M, Giudici-Orticoni MT, Lubitz W (2012). Chem. Communications, 2012, 48, 823-825.
-
[Fe4S4]- and [Fe3S4]-cluster formation in synthetic peptides. Hoppe A, Pandelia ME, Gärtner W, Lubitz W (2011). BBA-Bioenergetics 1807, 11,1414-1422.
-
Characterization of a unique [FeS] cluster in the electron transfer chain of the oxygen tolerant [NiFe] hydrogenase from Aquifex aeolicus. Pandelia ME, Nitschke W, Infossi P, Giudici-Orticoni MT, Bill E, Lubitz W (2011). Proc. Nat. Acad. Sci. USA 2011 Apr 12; 108(15): 6097–6102.
-
SEIRA spectroscopy of the electrochemical activation of an immobilized [NiFe] hydrogenase under turnover and non-turnover conditions. Millo D, Hildebrandt P, Pandelia ME, Lubitz W, Zebger I (2011). Ang. Chemie Int. Ed. 2011 Mar 7;50(11):2632-4.
-
The oxygen-tolerant hydrogenase I from Aquifex aeolicus weakly interacts with carbon monoxide: an electrochemical and time-resolved FTIR study. Pandelia ME, Infossi P, Giudici-Orticoni MT, Lubitz W (2010). Biochemistry. 2010 Oct 19;49(41):8873-81.
-
FTIR study on the light sensitivity of the [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F: Ni-C to Ni-L photoconversion, kinetics of proton rebinding and H/D isotope effect. Kellers P, Pandelia ME, Currell LJ, Görner H, Lubitz W (2009). Phys. Chem. Chem. Phys. 11: 8680 – 8683. *Equal authorship.
-
Membrane-bound hydrogenase I from the hyperthermophilic bacterium Aquifex aeolicus: enzyme activation, redox intermediates and oxygen tolerance. Pandelia ME, Fourmond V, Tron-Infossi P, Lojou E, Bertrand P, Léger C, Giudici-Orticoni MT, Lubitz W (2010). J Am Chem Soc. 2010 May 26;132(20):6991-7004.