Timothy Street
 Associate Professor of Biochemistry
Associate Professor of Biochemistry
Research Description
Mechanisms of protein folding in the cellThe balance of protein folding and degradation is one of the most fundamental activities of the cell, and is a critical point of intervention in cancer, metabolic, and aging diseases. Molecular chaperones are the central players that regulate the cell's repertoire of folded proteins, and as a consequence chaperones influence virtually every cellular process under both healthy and disease conditions. Despite their central influence, the functional mechanisms of many chaperones are poorly understood. Work in my lab is focused on revealing these mechanisms.
Dissecting a chaperone mechanism requires working at the interface of structural biology and protein folding. From a structural view, chaperones represent a fascinating class of molecular machines that undergo dramatic structural changes, often by utilizing ATP chemical energy (for example, see conformational changes of the Hsp90 chaperone in adjacent figure). These motions are then coupled to changes in the folding of their substrate proteins. However, it is not known how chaperones identify their binding partners in the complex cellular environment, and it is not known how chaperones affect protein folding.
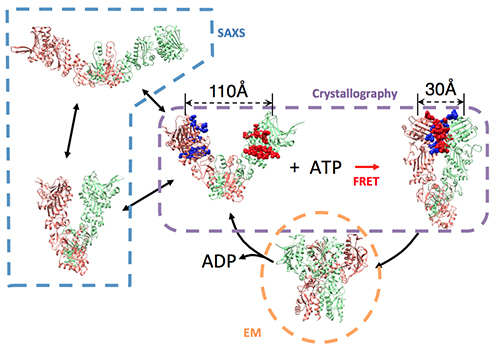
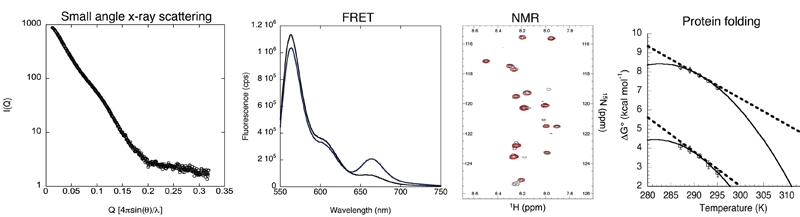
These fundamental questions will be answered by combining structural tools (x-ray crystallography, NMR, FRET, small angle x-ray scattering) with the tools of protein folding (thermodynamics, kinetics, energy landscape concepts) and computational modeling. In addition to these established techniques, new assays are critically needed to test chaperone mechanisms in live cells. One possibility is to design in-vivo protein-folding biosensors, which could have far-reaching practical applications.
Some therapeutic proteins are challenging to produce on a large scale because they require chaperones for optimal folding. There is a strong economic incentive to optimize the folding efficiency of these proteins. As a result, there is an exciting potential to use the mechanistic principles underlying chaperone function to rationally design new therapeutic proteins that fold more efficiently.
Selected Publications
- Jin Y, Kotler JLM, Wang S, Huang B, Halpin JC, Street TO (2021). The ER Chaperones BiP and Grp94 Regulate the Formation of Insulin-Like Growth Factor 2 (IGF2) Oligomers. J Mol Biol. 2021 Jun 25;433(13):166963. doi: 10.1016/j.jmb.2021.166963.
- Hoxie RS, Street TO (2020). Hsp90 chaperones have an energetic hot-spot for binding inhibitors. Protein Sci. 2020 Oct;29(10):2101-2111. doi: 10.1002/pro.3933.
- Halpin JC, Jangi R, Street TO (2020). Multimapping confounds ribosome profiling analysis: A case-study of the Hsp90 molecular chaperone. Proteins. 2020 Jan;88(1):57-68. doi: 10.1002/prot.25766.
- Huang B, Friedman LJ, Sun M, Gelles J, Street TO (2019). Conformational Cycling within the Closed State of Grp94, an Hsp90-Family Chaperone. J Mol Biol. 2019 Aug 9;431(17):3312-3323. doi: 10.1016/j.jmb.2019.06.004. Epub 2019 Jun 14.
- Sun M, Kotler JLM, Liu S, Street TO (2019). The endoplasmic reticulum (ER) chaperones BiP and Grp94 selectively associate when BiP is in the ADP conformation. J Biol Chem. 2019 Apr 19;294(16):6387-6396. doi: 10.1074/jbc.RA118.007050. Epub 2019 Feb 20.
-
Halpin JC, Street TO (2017). Hsp90 Sensitivity to ADP Reveals Hidden Regulation Mechanisms. J Mol Biol. 2017 Sep 15;429(19):2918-2930. doi: 10.1016/j.jmb.2017.08.005.
-
Jin Y, Hoxie RS, Street TO (2017). Molecular mechanism of bacterial Hsp90 pH-dependent ATPase activity. Protein Sci. 2017 Apr 6.
-
Liu S, Street TO. (2016) 5'-N-ethylcarboxamidoadenosine is not a paralog-specific Hsp90 inhibitor. Protein Sci. 2016 Dec;25(12):2209-2215.
-
Halpin JC, Huang B, Sun M and Street TO. (2016) Crowding Activates Heat Shock Protein 90. J Biol Chem 291(12): 6447-6455.
-
Turman DL, Nathanson JT, Stockbridge RB, Street TO, Miller C. (2015) Two-sided block of a dual-topology F- channel. Proc Natl Acad Sci USA. 2015 May 5;112(18):5697-701..
-
Street TO, Zeng X, Pellarin R, Bonomi M, Sali A, Kelly MJ, Chu F, Agard DA. (2014) Elucidating the mechanism of substrate recognition by the bacterial Hsp90 molecular chaperone. J Mol Biol. 2014 Jun 12;426(12):2393-404.
-
Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. (2013) Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol Cell. 2013 Feb 7;49(3):464-73.
-
Street TO, Lavery LA, Verba KA, Lee CT, Mayer MP, Agard DA. (2012) Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J Mol Biol. 2012 Jan 6;415(1):3-15.
-
Street TO, Lavery LA, Agard DA. (2011) Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011 Apr 8;42(1):96-105..
-
Krukenberg KA, Street TO, Lavery LA, Agard DA. (2011) Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys. 2011 May;44(2):229-55.
-
Street TO, Krukenberg KA, Rosgen J, Bolen DW, Agard DA. (2010) Osmolyte-induced conformational changes in the Hsp90 molecular chaperone. Protein Sci. 2010 Jan;19(1):57-65. doi: 10.1002/pro.282.
-
Krukenberg KA, Southworth DR, Street TO, Agard DA. (2009) pH-dependent conformational changes in bacterial Hsp90 reveal a Grp94-like conformation at pH 6 that is highly active in suppression of citrate synthase aggregation. J Mol Biol. 2009 Jul 10;390(2):278-91
-
Street TO, Barrick D. (2009) Predicting repeat protein folding kinetics from an experimentally determined folding energy landscape. Protein Sci. 2009 Jan;18(1):58-68. doi: 10.1002/pro.9.
-
Perskie LL, Street TO, Rose GD. (2008) Structures, basins, and energies: a deconstruction of the Protein Coil Library. Protein Sci. 2008 Jul;17(7):1151-61.
-
Street TO, Courtemanche N, Barrick D (2008). Protein folding and stability using denaturants. Methods Cell Biol. 2008;84:295-325.
-
Street TO, Fitzkee NC, Perskie LL, Rose GD (2007). Physical-chemical determinants of turn conformations in globular proteins. Protein Sci.2007 Aug;16(8):1720-7.
-
Street TO, Bradley CM, Barrick D (2007). Predicting coupling limits from an experimentally determined energy landscape. Proc Natl Acad Sci U S A. 2007 Mar 20;104(12):4907-12.
-
Street TO, Bolen DW, Rose GD (2006). A molecular mechanism for osmolyte-induced protein stability. Proc Natl Acad Sci U S A. 2006 Sep 19;103(38):13997-4002.
-
Street TO, Rose GD, Barrick D (2006). The role of introns in repeat protein gene formation. J Mol Biol. 2006 Jul 7;360(2):258-66.
-
Street TO, Bradley CM, Barrick D (2005). An improved experimental system for determining small folding entropy changes resulting from proline to alanine substitutions. Protein Sci. 2005 Sep;14(9):2429-35.
-
Fitzkee NC, Fleming PJ, Gong H, Panasik N Jr, Street TO, Rose GD (2005). Are proteins made from a limited parts list? Trends Biochem Sci.2005 Feb;30(2):73-80.
-
Sun M, Kotler JLM, Liu S, Street TO (2019). The ER chaperones BiP and Grp94 selectively associate when BiP is in the ADP conformation. J Biol Chem. 2019 Feb 20. pii: jbc.RA118.007050. doi: 10.1074/jbc.RA118.007050.
-
Halpin JC, Street TO. Hsp90 Sensitivity to ADP Reveals Hidden Regulation Mechanisms. J Mol Biol. 2017 Sep 15;429(19):2918-2930. doi: 10.1016/j.jmb.2017.08.005.