Alexandre Bisson
 Assistant Professor of Biology
Assistant Professor of BiologyResearch Description
Cellular Organization and Behavior in the Archaea domain of life
How the evolution of molecular systems shaped the function and behavior of cells and created the life diversity observed today is still an exciting puzzle. From the apparent simplicity of bacterial cells to the multi-compartmentalized eukaryotes, all these organisms are capable of employing similar molecular components by different mechanisms through different physical and temporal scales to propagate and survive. To understand how such complexity emerged and the mechanisms in which is regulated, we aim to study the cell biology of Archaea.
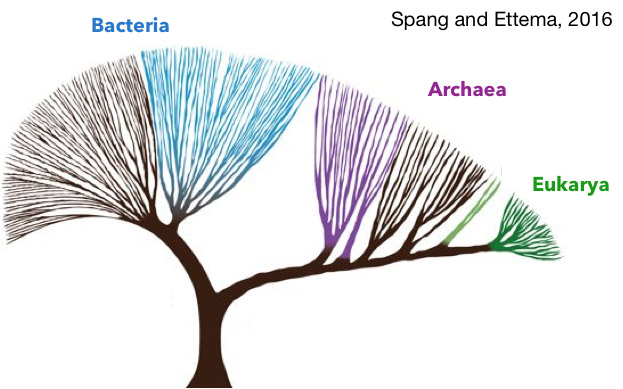
Low-resolution tree of life representing the distribution and divergence of the 3 domains of life. Adapted from Spang and Ettema, 2016.
Archaea (from Greek arkhaios, ‘primitive’) are unique microorganisms that equipped with genes previously thought to be present exclusive of bacteria or eukaryotes, summoning an exciting hybrid of molecular components that not only predated eukaryotes but might have played an essential role in their emergence.
The billions of years separating Bacteria and Eukaryotes qualify archaeal cell biology as one of the most promising fields for unseen biological phenomena.
To break new ground, the Bisson Lab is pioneering in a variety of orthogonal approaches: 1) plasmid-free genetic engineering and genetic screens to identify new cellular components; 2) advanced live-cell microscopy and computational analysis to observe and quantitate the in vivo dynamics of molecular components; 3) in vitro biochemical reconstitutions to elucidate their kinetic parameters; 4) Microfabrication and cultivation of complex microbial populations within microfluidic environments.
In our group, intellectual independence and freedom to pursue one's curiosity are fundamental assets. That means that we welcome ideas for new topics that are not currently ongoing in the lab. However, few questions permeate our projects and guide our research:
- What are the mechanisms that control archaeal cell shape and how to reproduce such diversity in vitro and other organisms?
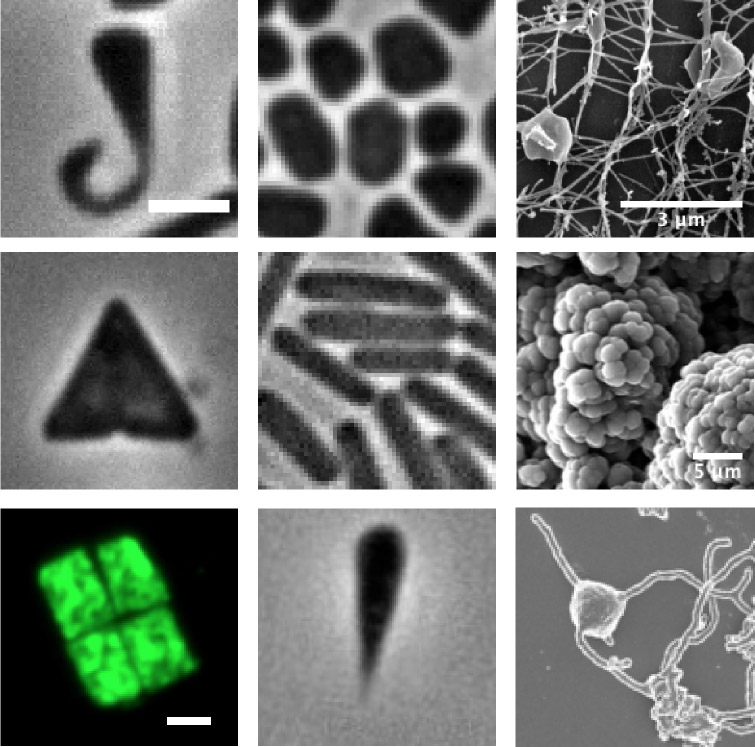
Shape diversity across different archaea. H. walsbyi image kindly provided by Mike Dyall-Smith (U. of Melbourne). Images from third column were adapted from Reiger, 1995 (top), Conklin et al., 2006 (center) and Imachi et al., 2019 (bottom). Unless otherwise stated, scale bars represents 2 μm.
- How do archaea regulate cell division and pace their cell cycle?
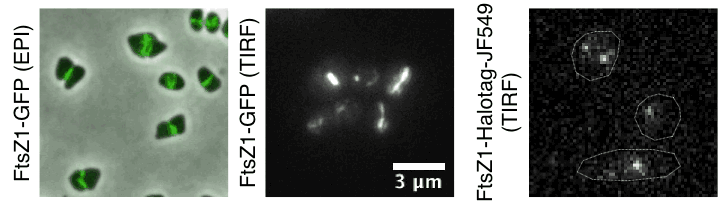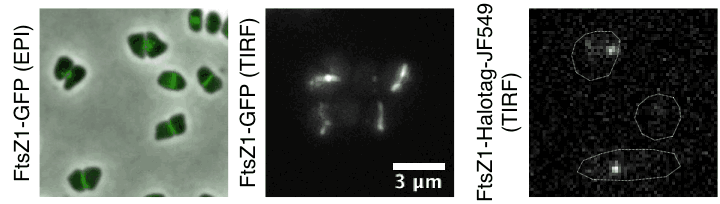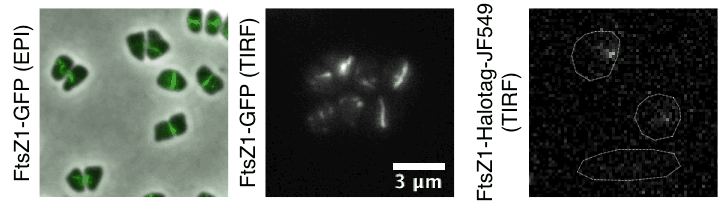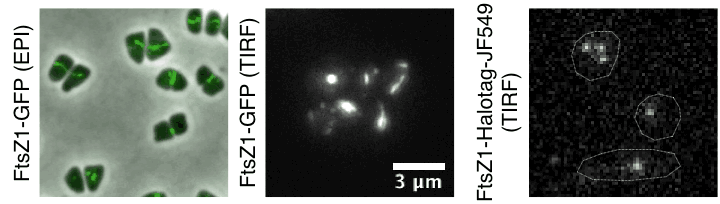
Time lapse of haloarchaeal cytoskeleton dynamics during cell division in Haloarchaea. Movies show unusual cytokinesis directional motion of tubulin-like filaments and their single-molecule dynamics
- How do archaea sense the physical and chemical environment around them?
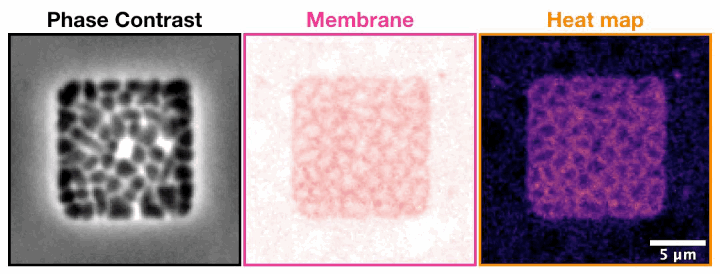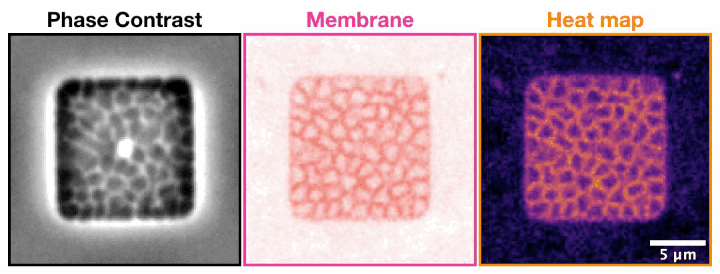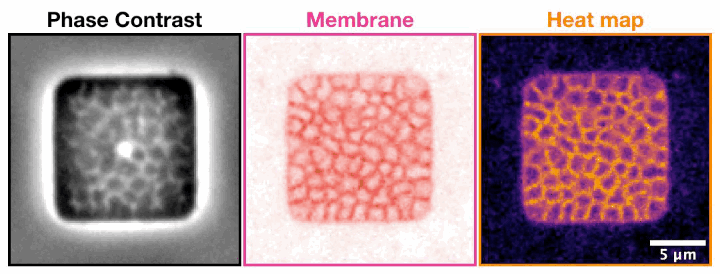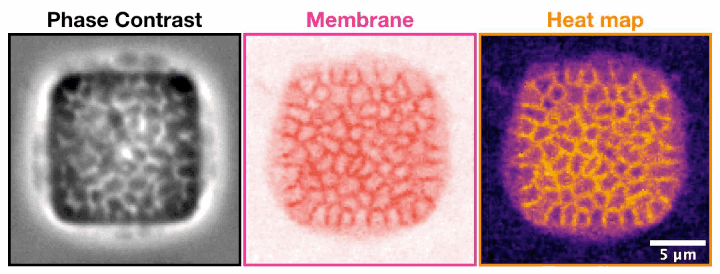
Archaeal cells trapped inside microfabricated chambers back” compression by decreasing membrane fluidity.
Publication List
Here is a list of our papers in the past 5 years.
- Lipid Anchoring of Archaeosortase Substrates and Midcell Growth in Haloarchaea. Abdul-Halim MF, Schulze S, DiLucido A, Pfeiffer F, Bisson Filho AW, Pohlschroder M. mBio. 2020 Mar 24;11(2):e00349-20. doi: 10.1128/mBio.00349-20.
- A survey-based analysis of the academic job market. Fernandes JD, Sarabipour S, Smith CT, Niemi NM, Jadavji NM, Kozik AJ, Holehouse AS, Pejaver V, Symmons O, Bisson Filho AW, Haage A. Elife. 2020 Jun 12;9:e54097. doi: 10.7554/eLife.54097.
- The Ribbon-Helix-Helix Domain Protein CdrS Regulates the Tubulin Homolog ftsZ2 To Control Cell Division in Archaea. Darnell CL, Zheng J, Wilson S, Bertoli RM, Bisson-Filho AW, Garner EC, Schmid AK. mBio. 2020 Aug 11;11(4):e01007-20.
- Haloferax volcanii Immersed Liquid Biofilms Develop Independently of Known Biofilm Machineries and Exhibit Rapid Honeycomb Pattern Formation. Schiller H, Schulze S, Mutan Z, de Vaulx C, Runcie C, Schwartz J, Rados T, Bisson Filho AW, Pohlschroder M. mSphere. 2020 Dec 16;5(6):e00976-20. doi: 10.1128/mSphere.00976-20.
- Walsh, J. C., Angstmann, C. N., Bisson‐Filho, A. W., Garner, E. C., Duggin, I. G. and Curmi, P. M. (2019), Division plane placement in pleomorphic archaea is dynamically coupled to cell shape. Mol Microbiol. 2019 112:3, 785-799.
- Alexandre W. Bisson-Filho, Jenny Zheng, and Ethan Garner. (2018), Archaeal imaging: leading the hunt for new discoveries. Molecular Biology of the Cell 2018 29:14, 1675-1681.
- Saman Hussain, Carl N Wivagg, Piotr Szwedziak, Felix Wong, Kaitlin Schaefer, Thierry Izoré, Lars D Renner, Matthew J Holmes, Yingjie Sun, Alexandre W Bisson-Filho, Suzanne Walker, Ariel Amir, Jan Löwe, Ethan C Garner. (2018), MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 2018;7:e32471.
- Alexandre W Bisson-Filho, Yen-Pang Hsu, Georgia R Squyres, Erkin Kuru, Fabai Wu, Calum Jukes, Yingjie Sun, Cees Dekker, Seamus Holden, Michael S VanNieuwenhze, Yves V Brun, Ethan C Garner. (2017), Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science. 355(6326):739-743.
- Diego Emiliano Sastre, Alexandre Bisson‐Filho, Diego de Mendoza, Frederico J Gueiros‐Filho. (2016), Revisiting the cell biology of the acyl‐ACP:phosphate transacylase PlsX suggests that the phospholipid synthesis and cell division machineries are not coupled in Bacillus subtilis. Molecular Microbiology, 100: 621-634.