Chandler Fulton
 Professor Emeritus of Biology
Professor Emeritus of Biology
Research Description
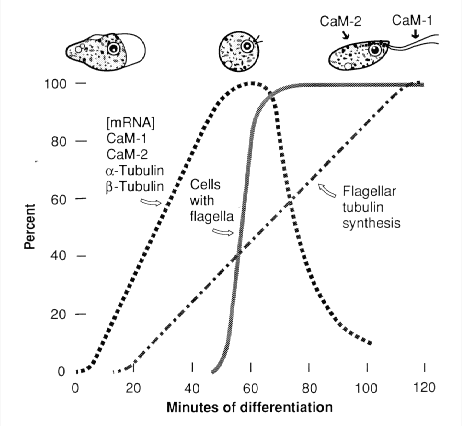
In the 1960s my lab set out to use genetics to dissect cell differentiation. We chose as research organism a remarkable unicellular eukaryote that lives in freshwater, Naegleria gruberi, which can alternate between walking amoebae and swimming flagellates. When amoebae are transferred from their growth environment to a nutrient-free aqueous environment, they undergo synchronous differentiation to streamlined flagellates, a conversion completed within 60 to 100 minutes. We found that these differentiating cells undergo a yin-yang change in utilization of two eukaryotic motility systems. Amoebae walk using an actin-based motility system, which becomes latent in the flagellates, and the actin mRNA of amoebae is rapidly destroyed during differentiation. The flagellates swim using a tubulin-based motility system, but the tubulin that makes up the flagellar and basal body microtubules is entirely synthesized as a programmed event of differentiation. Evidence that this differentiation is regulated in part by changes in intracellular free calcium ions led us to calmodulin, the conserved calcium-binding protein that in all eukaryotes transduces the calcium signal to diverse effector proteins. Two calmodulins were found in Naegleria flagellates, and they are neatly segregated in the cells, with the major one (CaM-1) localized in the flagella and the other (CaM-2) in the flagellate cell body. During differentiation the mRNAs for the two calmodulins increase in abundance and then rapidly decrease concurrently with those for α- and β-tubulins (Fulton et al., 1995). We also have studied the formation of the centriolar 9-triplet basal bodies, and have shown that the two basal bodies of flagellates form in a two-step process by self-assembly of the first and then mentored assembly of the second basal body. In collaboration with Lillian Fritz-Laylin, we have shown that these two modes of centriole assembly are ancient and have been conserved through the evolution of eukaryotes.
Cell Differentiation in Naegleria gruberi
Dissection of the signal transduction pathways from initiation of Naegleria differentiation through the regulation of transcription, mRNA stability, and localization of proteins in the flagella depends on genetics, but in spite of much effort N. gruberi has always behaved as if asexual in the lab, and in addition has so far resisted our efforts to utilize the powerful techniques of molecular genetics. Yet when we collaboratively sequenced the genome of the clonal strain we study (Fritz-Laylin et al., 2010), we found that this strain is a diploid heterozygote from a mating that occurred in nature more than a half-century ago. In addition, the strain still retains among its 16,000 genes functional genetic toolkits for meiosis and mating. We continue to seek conditions that would induce N. gruberi to form gametes and mate in the laboratory, but in the meantime have been approaching the challenge of achieving genetics in other ways. After a broad survey of the ~40 species of Naegleria in collaboration with Larry Wangh’s lab, we have focused on one species, N. minor, which appears to form gametes and to mate under lab conditions, and we are studying that process.
Our efforts to manipulate genes of N. gruberi using DNA-mediated transformation led us to the surprising discovery that successful transformation, even with a wild-type gene, causes the chromosomal gene to be silenced. The silencing involves no change in the base sequence of the gene, and is stable for generations, but is metastable in that the gene eventually regains its activity. In an ongoing collaborative study with New England Biolabs, we are working to understand how this epigenetic silencing works, and to learn if this mechanism of gene silencing might have broader utility.
We found unexpectedly that Naegleria contains an agent that induces apoptosis in vertebrate cells. The Naegleria agent, a small protein, is not toxic to vertebrate cells and does not affect their growth, but causes them to die in a delayed fashion after the cells exit the cell cycle. It induces apoptosis independently of oncogene mutations that render many chemotherapeutic agents ineffectual. We have identified this agent as a thiaminase, which produces delayed apoptosis by causing a deficiency in vitamin B1 (see Elaine Lai’s webpage). This agent offers unique opportunities for targeted cancer therapy, and we hope to find partners to explore possible therapeutic uses.
Selected Publications
- Fritz-Laylin LK and Fulton C (2016). "Naegleria: a classic model for de novo basal body assembly." Cilia 2016 Apr 4;5:10.
- Fritz-Laylin LK, Levy YY, Levitan E, Chen S, Cande WZ, Lai EY and Fulton C (2016). "Rapid centriole assembly in Naegleria reveals conserved roles for both de novo and mentored assembly." Cytoskeleton (Hoboken) 73(3): 109-116.
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC (2010). "The genome of Naegleria gruberi illuminates early eukaryotic versatility." Cell. 2010 140:631-42.
- Levy YY, Lai EY, Remillard SP, Fulton C. (1998). "Centrin is synthesized and assembled into basal bodies during Naegleriadifferentiation." Cell Motil Cytoskeleton. 40:249-60.
- Fulton C, Lai EY, Remillard SP (1995). "A flagellar calmodulin gene of Naegleria, coexpressed during differentiation with flagellar tubulin genes, shares DNA, RNA, and encoded protein sequence elements." J Biol Chem. 1995 Mar 17;270:5839-48.
- Fulton C (1993). "Naegleria: A research partner for cell and developmental biology." J. Euk. Microbiol. 40:520-32.
- Fulton C (1970). "Amebo-flagellates as research partners: The laboratory biology of Naegleria and Tetramitus." Methods Cell Physiol. (D. M. Prescott, ed.). 4:341-476.