Bruce Goode
 Professor of Biology
Professor of Biology
Department Chair, Biology
Research Description
Cytoskeletal Assembly and Dynamics
Our lab is focused on the question of how cells dynamically assemble and reorganize their actin cytoskeletons to govern cell shape, cell movement, and cell division. Every cell type has a unique architecture tailored to its physiological functions, which is defined in large part by its cytoskeleton. The cytoskeleton is a vast system of interconnected polymeric tubes and fibers that creates a dynamic scaffold used to produce polarity and compartmentalization, and to generate force. The primary aim of our research is to understand how cells bring about rapid and precise rearrangements of their actin polymer networks, transforming cell shape and function.
Actin filament networks are complex, self-assembling biological machines built from hundreds of distinct components and moving parts. We have been dissecting the inner workings of these force-producing machines, analyzing the cellular functions and biochemical mechanisms of actin regulatory proteins in order to understand how this machinery collectively orchestrates the assembly, reorganization, and disassembly of entire actin networks. Our goal is to obtain a highly mechanistic and quantitative view of these events.
We use two different in vivo systems: (a) the budding yeast S. cerevisiae, which offers advanced genetic analyses; and (b) mammalian fibroblasts and primary cardiomyocytes, which offer superior cytology and enable us to study actin regulation underlying cell motility and muscle contraction. Two fundamental questions are being addressed in the lab:
- How is the rapid assembly and disassembly of actin networks governed? To answer this question, we are studying the roles of crucial actin regulatory proteins that alter polymer dynamics, e.g. formins, Arp2/3 complex, and ADF/cofilin (figure 1). We have also identified novel binding partners of these regulators that have surprising new activities. This work includes the characterization of: (a) yeast and mammalian formins, (b) formin-binding proteins (e.g. APC, Bud6, Bud14 and Smy1), (c) WASp-Arp2/3 complex binding partners (e.g. Abp1, Syp1, coronin, and GMF), and (d) the actin disassembly and turnover machinery (e.g. Aip1, coronin, twinfilin, and Srv2/CAP).
- How is the regulation of actin dynamics coordinated with regulation of other cytoskeletal polymer systems (microtubules, septins, and intermediate filaments)? Our recent work in this area addresses functional cross-talk between the actin and microtubule cytoskeletons regulated by a triad of mammalian proteins that physically associate: (a) the tumor suppressor protein Adenomatous polyposis coli (APC), (b) the formin Dia1, and (c) the microtubule end-binding protein EB.
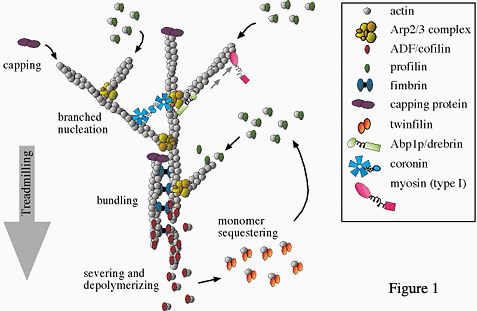
Figure 1. Crucial actin regulatory proteins that alter polymer dynamics, e.g. formins, Arp2/3 complex, and ADF/cofilin.
To answer the questions above, we are taking a multidisciplinary approach combining genetics, biochemistry, structural biology, and live cell imaging. In vitro analyses include quantitative fluorescence-based kinetic assays, time lapse TIRF microscopy on individual actin filaments, single molecule analysis on actin regulatory proteins, and reconstituted actin-based bead motility assays in cell extracts and purified systems. In vivo analyses include forward and reverse genetic screens, RNAi silencing, live-cell imaging to track cytoskeletal regulatory protein dynamics and polymer dynamics, and electron microscopy (figure 2).

Figure 2. In vivo analyses include forward and reverse genetic screens, RNAi silencing, live-cell imaging to track cytoskeletal regulatory protein dynamics and polymer dynamics, and electron microscopy
Selected Publications
View Complete Publication List on PubMed: Bruce Goode
- DAAM2 Variants Cause Nephrotic Syndrome via Actin Dysregulation. Schneider R, Deutsch K, Hoeprich GJ, Marquez J, Hermle T, Braun DA, Seltzsam S, Kitzler TM, Mao Y, Buerger F, Majmundar AJ, Onuchic-Whitford AC, Kolvenbach CM, Schierbaum L, Schneider S, Halawi AA, Nakayama M, Mann N, Connaughton DM, Klämbt V, Wagner M, Riedhammer KM, Renders L, Katsura Y, Thumkeo D, Soliman NA, Mane S, Lifton RP, Shril S, Khokha MK, Hoefele J, Goode BL, Hildebrandt F. Am J Hum Genet. 2020 Dec 3;107(6):1113-1128.
- Single-molecule imaging of IQGAP1 regulating actin filament dynamics. Hoeprich GJ, Sinclair AN, Shekhar S, Goode BL. Mol Biol Cell. 2022 Jan 1;33(1):ar2. doi: 10.1091/mbc.E21-04-0211.
- Scaling of subcellular actin structures with cell length through decelerated growth. McInally SG, Kondev J, Goode BL. Elife. 2021 Jun 11;10:e68424. doi: 10.7554/eLife.68424.
- Bil2 Is a Novel Inhibitor of the Yeast Formin Bnr1 Required for Proper Actin Cable Organization and Polarized Secretion. Rands TJ, Goode BL. Front Cell Dev Biol. 2021 Feb 9;9:634587. doi: 10.3389/fcell.2021.634587. eCollection 2021.
- Twinfilin bypasses assembly conditions and actin filament aging to drive barbed end depolymerization. Shekhar S, Hoeprich GJ, Gelles J, Goode BL. J Cell Biol. 2021 Jan 4;220(1):e202006022. doi: 10.1083/jcb.202006022.
- EB1 Directly Regulates APC-Mediated Actin Nucleation. Juanes MA, Fees CP, Hoeprich GJ, Jaiswal R, Goode BL. Curr Biol. 2020 Dec 7;30(23):4763-4772.e8. doi: 10.1016/j.cub.2020.08.094.
- Cell-substrate adhesion drives Scar/WAVE activation and phosphorylation by a Ste20-family kinase, which controls pseudopod lifetime. Singh SP, Thomason PA, Lilla S, Schaks M, Tang Q, Goode BL, Machesky LM, Rottner K, Insall RH. PLoS Biol. 2020 Aug 3;18(8):e3000774. doi: 10.1371/journal.pbio.3000774. eCollection 2020 Aug.
- Cofilin Loss in Drosophila Muscles Contributes to Muscle Weakness through Defective Sarcomerogenesis during Muscle Growth. Balakrishnan M, Yu SF, Chin SM, Soffar DB, Windner SE, Goode BL, Baylies MK. Cell Rep. 2020 Jul 21;32(3):107893. doi: 10.1016/j.celrep.2020.107893.
- WAVE1 and WAVE2 have distinct and overlapping roles in controlling actin assembly at the leading edge. Tang Q, Schaks M, Koundinya N, Yang C, Pollard LW, Svitkina TM, Rottner K, Goode BL. Mol Biol Cell. 2020 Sep 15;31(20):2168-2178. doi: 10.1091/mbc.E19-12-0705.
- A septin-Hof1 scaffold at the yeast bud neck binds and organizes actin cables. Garabedian MV, Wirshing A, Vakhrusheva A, Turegun B, Sokolova OS, Goode BL. Mol Biol Cell. 2020 Aug 15;31(18):1988-2001. doi: 10.1091/mbc.E19-12-0693.
- Centering and symmetry breaking in confined contracting actomyosin networks. Ierushalmi N, Malik-Garbi M, Manhart A, Abu Shah E, Goode BL, Mogilner A, Keren K. Elife. 2020 Apr 21;9:e55368. doi: 10.7554/eLife.55368.
- Genetically inspired in vitro reconstitution of Saccharomyces cerevisiae actin cables from seven purified proteins. Pollard LW, Garabedian MV, Alioto SL, Shekhar S, Goode BL. Mol Biol Cell. 2020 Mar 1;31(5):335-347. doi: 10.1091/mbc.E19-10-0576.
- Synergy between Cyclase-associated protein and Cofilin accelerates actin filament depolymerization by two orders of magnitude. Shekhar S, Chung J, Kondev J, Gelles J and Goode BL. Nature Communications. 2019 Nov 22;10(1):5319. doi: 10.1038/s41467-019-13268-1.
- Scaling behavior in steady-state contracting actomyosin networks. Malik-Garbi M, Ierushalmi N, Jansen S, Abu-Shah E, Goode BL, Mogilner A, Keren K. Nature Physics. 2019 May;15(5):509-516. doi: 10.1038/s41567-018-0413-4.
- The role of APC-mediated actin assembly in microtubule capture and focal adhesion turnover. Juanes MA, Isnardon D, Badache A, Brasselet S, Mavrakis M, Goode BL. Journal of Cell Biology. 2019 Oct 7;218(10):3415-3435. doi: 10.1083/jcb.201904165.
- Tropomyosin Isoforms differentially tune actin filament length and disassembly. Jansen S, Goode BL. Mol. Biol. Cell. 2019 Mar 1;30(5):671-679. doi: 10.1091/mbc.E18-12-0815.
- A novel mode of Capping Protein-regulation by Twinfilin. Johnston AB, Hilton DM, McConnell P, Johnson B, Harris MT, Simone A, Amarasinghe GK, Cooper JA, Goode BL. eLife 2018;7:e41313.
- Integrated control of formin-mediated actin assembly by a stationary inhibitor and a mobile activator. Garabedian MV, Stanishneva-Konovalova T, Lou C, Rands TJ, Pollard LW, Sokolova OS, Goode BL. Journal of Cell Biology. Oct 1;217(10):3512-3530.
- Abp1 promotes Arp2/3 complex-dependent actin nucleation and stabilizes branch junctions by antagonizing GMF. Guo S, Sokolova OS, Chung J, Padrick S, Gelles J, Goode BL (2018). Nature Communications Jul 24;9(1):2895.
- Species-specific Functions of Twinfilin in Actin Filament Depolymerization. Hilton DM, Aguilar RM, Johnston AB, and Goode BL (2018). Journal of Molecular Biology Sep 14;430(18 Pt B):3323-3336.
- GMF as an Actin Network Remodeling Factor. Goode BL, Sweeney MO, Eskin JA. Trends Cell Biol. May 18. pii: S0962-8924(18)30072-2.
- Structural basis of actin monomer re-charging by cyclase-associated protein. Kotila T, Kogan K, Enkavi G, Guo S, Vattulainen I, Goode BL, Lappalainen P. Nature Communications May 14;9(1):1892. doi: 10.1038/s41467-018-04231-7.
- Rapid Production of Pure Recombinant Actin Isoforms in Pichia pastoris. Hatano T, Alioto S, Roscioli E, Palani S, Clarke ST, Kamnev A, Hernandez-Fernaud JR, Lavanya Sivashanmugam, Bernardo Chapa-Y-Lazo, Alexandra Jones, Robinson R, Sampath K, Mishima M, McAinsh AD, Goode BL, Balasubramaniam M. J Cell Sci. 2018 Apr 15; 131(8): jcs213827.
- A Flow Cytometry-Based Phenotypic Screen To Identify Novel Endocytic Factors in Saccharomyces cerevisiae. Wrasman K, Alioto S, Zhang Y, Hoban K, Khairy M, Goode BL, Wendland B. G3 (Bethesda). 2018 May; 8(5): 1497–1512.
- Sokolova OS, Chemeris A, Guo S, Alioto SL, Gandhi M, Padrick S, Pechnikova E, David V, Gautreau A and Goode BL (2017). Structural Basis of Arp2/3 Complex Inhibition by GMF, Coronin, and Arpin. J Mol Biol 429(2): 237-248.
- Alioto SL, Garabedian MV, Bellavance DR, Goode BL (2016). Tropomyosin and Profilin Cooperate to Promote Formin-Mediated Actin Nucleation and Drive Yeast Actin Cable Assembly. Curr Biol. 26:3230–3237, 5 December 2016
- Mohapatra L, Goode BL, Jelenkovic P, Phillips R, Kondev J. (2016). Design Principles of Length Control of Cytoskeletal Structures. Annual Review of Biophysics, 42:85-116.
- Henty-Ridilla J, Rankova A, Eskin JA, Kinney K and Goode BL (2016). Accelerated actin polymerization from microtubule plus ends. Science. 352(6288):1004-9. doi: 10.1126/science.aaf1709.
- Chin SM, Jansen S, Goode BL (2016). TIRF microscopy analysis of human Cof1, Cof2, and ADF effects on actin filament severing and turnover. J. Mol. Biol. 428:1604-1616.
- Eskin JA, Rankova A, Johnston AB, Alioto SA, and Goode BL (2016). Common formin-regulating sequences in Smy1 and Bud14 are required for the control of actin cable assembly in vivo. Mol. Biol. Cell. 5:828-837.
- Bombardier JP, Eskin JA, Jaiswal R, Correa IR Jr., Xu MQ, Goode BL, Gelles J (2015). Single-molecule visualization of a formin-capping protein "decision complex" at the actin filament barbed end. Nature Communications 6: 8707.
- Boopathy S, Silvas TV, Tischbein M, Jansen S, Shandilya SM, Zitzewitz JA, Landers JE, Goode BL, Schiffer CA, Bosco DA (2015). Structural basis for mutation-induced destabilization of profilin 1 in ALS. Proc Natl Acad Sci U S A 112(26): 7984-7989.
- Goode BL, Eskin JA, Wendland B. (2015). Actin and endocytosis in budding yeast. Genetics. 2015 Feb;199(2):315-58.
- Henty-Ridilla JL and Goode BL (2015). Global resource distribution: allocation of actin building blocks by profilin. Dev Cell. 2015 Jan 12;32(1):5-6.
- Jansen S, Collins A, Chin SM, Ydenberg CA, Gelles J, Goode BL (2015). Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nature Communications 2015 May 21;6:7202.
- Jansen S, Collins A, Golden L, Sokolova O, Goode BL (2014). Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J Biol Chem 289(44): 30732-30742.
- Johnston AB, Collins A, Goode BL (2015). High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat Cell Biol 17(11): 1504-1511.
- Mikati MA, Breitsprecher D, Jansen S, Reisler E, Goode BL (2015). Coronin Enhances Actin Filament Severing by Recruiting Cofilin to Filament Sides and Altering F-Actin Conformation. J Mol Biol 427(19): 3137-3147.
- Mohapatra L, Goode BL, Kondev J (2015). Antenna Mechanism of Length Control of Actin Cables. PLoS Comput Biol. 2015 Jun 24;11(6):e1004160.
- Park E, Graziano BR, Zheng W, Garabedian M, Goode BL, Eck MJ (2015). Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif. Structure 23(8): 1492-1499.
- Poukkula M, Hakala M, Pentinmikko N, Sweeney MO, Jansen S, Mattila J, Hietakangas V, Goode BL, Lappalainen P (2014). GMF promotes leading-edge dynamics and collective cell migration in vivo. Curr Biol. 2014 Nov 3;24(21):2533-40.
- Sweeney MO, Collins A, Padrick SB, Goode BL (2015). A novel role for WAVE1 in controlling actin network growth rate and architecture. Mol Biol Cell. 2015 Feb 1;26(3):495-505.
- Ydenberg CA, Johnston A, Weinstein J, Bellavance D, Jansen S, Goode BL (2015). Combinatorial genetic analysis of a network of actin disassembly-promoting factors. Cytoskeleton (Hoboken) 72(7): 349-361.
- Daou P, Hasan S, Breitsprecher D, Baudelet E, Camoin L, Audebert S, Goode BL, Badache A. (2014). Essential and nonredundant roles for Diaphanous formins in cortical microtubule capture and directed cell migration. Mol Biol Cell. 2014 Mar;25(5):658-68.
- Gould CJ, Chesarone-Cataldo M, Alioto SL, Salin B, Sagot I, and Goode BL (2014). S. cerevisiae Kelch proteins and Bud14 form a stable 520 kDa formin-regulatory complex that controls actin cable assembly and cell morphogenesis. J. Biol. Chem. 2014 Jun 27;289(26):18290-301.
- Graziano BR, Yu HY, Alioto SL, Eskin JA, Ydenberg CA, Waterman DP, Garabedian M, and Goode BL (2014). The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell. 2014 Jun 1;25(11):1730-43.
- Smith BA, Gelles J, and Goode BL (2014). Single-molecule studies of actin assembly and disassembly factors. Meth Enz. 2014; 540:95-117.
- Chaudhry F, Jansen S, Little K, Suarez C, Boujemaa-Paterski R, Blanchoin L, and Goode BL (2014). Autonomous and in trans functions for the two halves of Srv2/CAP in promoting actin turnover. Cytoskeleton (Hoboken). 2014 Jun;71(6):351-60.
- Rosado M, Barber CF, Berciu C, Feldman S, Birren SJ, Nicastro D, and Goode BL (2014). Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell. 2014 Mar;25(6):811-27.
- Jaiswal R, Breitsprecher D, Collins A, Correa IR, Xu MQ, and Goode BL (2013). The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr. Biol. 2013 Jul 22; 23:1373-1379.
- Ydenberg CA, Padrick SB, Sweeney MO, Gandhi M, Sokolova O, and Goode BL (2013). GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr Biol. 2013 Jun 17;23(12):1037-45.
- Graziano BR, Jonasson EM, Pullen JG, Gould CJ, and Goode BL (2013). Ligand-induced activation of a formin-NPF pair leads to collaborative actin nucleation. J Cell Biol. 2013 May 13;201(4):595-611.
- Breitsprecher D and Goode BL (2013). Formins at a glance. J Cell Sci. 2013 Jan 1;126:1-7.
- Chaudhry F, Breitsprecher D, Little K, Sharov G, Sokolova O, and Goode BL (2013). Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol Biol Cell. 2013 Jan;24:31-41.
- Maiti S, Michelot A, Gould CJ, Blanchoin L, Sokolova O, and Goode BL (2012). Structure and activity of full-length formin mDia1. Cytoskeleton. 2012 Jun;69:393-4055.
- Breitsprecher D, Jaiswal R, Bombardier J, Gould CJ, Gelles J, and Goode BL (2012). Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012 Jun 1;336:1164-1168.