James Haber
 Abraham and Etta Goodman Professor of Biology, and Director, Rosenstiel Basic Medical Sciences Research Center
Abraham and Etta Goodman Professor of Biology, and Director, Rosenstiel Basic Medical Sciences Research Center
Research Description
Repair of broken chromosomes and triggering of the DNA damage response
View lectures from James Haber
Broken chromosomes must be repaired if a cell is to survive; consequently cells have evolved a variety of mechanisms to repair double-strand breaks (DSBs). Both homologous recombination, in which the ends of the broken DNA seek out intact templates with the same sequence, and nonhomologous end-joining pathways are found in Saccharomyces as they are in humans. In addition cells have evolved a damage-sensing checkpoint system whereby the cells delay entry into mitosis until the break has been repaired.
Analysis of homologous recombination.
Recombination between homologous sequences is a fundamentally important process both in meiosis and in mitotic cells. We are interested in understanding at the molecular level how recombination occurs and what roles are played by the many proteins involved in DNA recombination, repair and replication. Using synchronized cells undergoing recombination that is initiated at a specific site on a chromosome by an inducible endonuclease, we use physical monitoring techniques (Southern blots, PCR analysis) to follow the sequence of molecular events that occur in real time. We are interested in determining what are the specific biochemical roles played by the many proteins implicated in DNA recombination, repair and replication. This "in vivo biochemistry" approach has enabled us to demonstrate that there are in fact several independent, competing pathways of homologous recombination, each with its own genetic requirements.
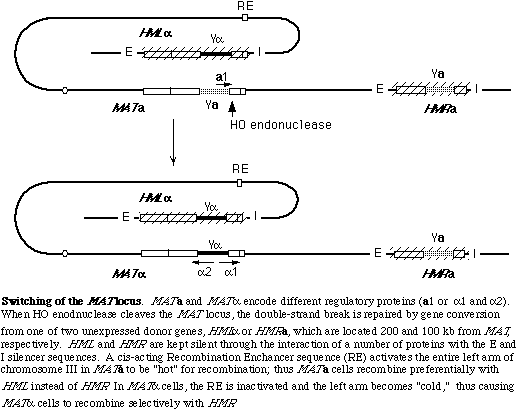
We have focused a lot of attention on yeast mating-type gene switching (MAT switching) as an example of DSB repair that we can study in great detail. Here a DSB at the MAT locus is created by a site specific HO endonuclease, which we can induce synchronously in a large population of cells. The DSB is repaired from one of two heterochromatic donors (HML and HMR). We have identified the proteins necessary to carry out the initial steps in strand invasion and the beginning of new DNA synthesis, which is significantly different from the normal process of replication. We have shown that the invasion of DNA strands into a donor template region requires the action of the chromatin remodeling protein Rad54 that enables the recombination machinery to gain access to "closed" regions of DNA. Recently we used a modification of this system to study the mutation rate associated with DSB repair. We find that there is a 1000-fold increase in mutation rate and that about half the mutations have a distinctive “signature” suggesting that the DNA polymerase frequently dissociates and re-anneals with its template.
MAT switching is an example of a repair process called gene conversion. This is the process we have studied most intensively, but we are also interested in an alternative process known as break-induced replication (BIR), where only one end of a chromosome break locates a template sequence and assembles a complete replication fork and can copy sequences to the end of a chromosome, producing a nonreciprocal translocation. We have shown that there are differences between this repair-induced replication fork and the normal replication process, but there is much more work to do.
We are also interested in gene targeting methods and in figuring out why these types of gene replacement and modification are quite inefficient, even in yeast. Finally we are interested in comparing how recombination occurs in mitosis and in meiosis. To this end we have expressed the site-specific HO endonuclease in meiotic cells so that we can compare recombination events at the same loci where we have used HO to stimulate recombination in mitotic cells.
Donor preference
We have been fascinated by the process of yeast mating-type gene switching, in which cells replace about 700 bp of Ya or Y-specific DNA sequences at the MAT locus by recombining with one of two donor loci, called HMLDescription: image3 and HMRa. The two donor loci are maintained in a chromatin configuration that prevents them from being transcribed or being cleaved by the HO endonuclease that cuts the same sequence at MAT to initiate switching. In addition to determining how this process occurs and how various mutations affect it, we are particularly interested in the phenomenon of donor preference, whereby MATa cells choose the donor on the left while MAT elects to recombine with the donor on the right, even if we replace HML by HMR; it is the position on the chromosome that dictates donor choice. We have shown that this regulation involves the action of a small Recombination Enhancer (RE) sequence that enables a donor on the left chromosome arm to recombine preferentially in MATa cells. This element is turned off in MAT cells, so the HMR donor on the right is preferred. Recently we have shown that RE binds multiple copies of a transcription factor, Fkh1. Further work has shown that only a part of Fkh1 – the FHA domain that binds a particular phosphorylated threonine – is required. We have recently shown that RE directly interacts with the site of a DSB, and that in so doing it pulls HML close to the break. Now we need to figure out what is the specific target of FHA domain.
Nonhomologous End-Joining and Repair.
In addition to repair of a double-strand chromosomal break by homologous recombination mechanisms, we have also demonstrated that yeast--like mammalian cells--also employ several nonhomologous repair pathways. These different pathways have distinct cell cycle and genetic requirements.
We are also interested in the addition of new telomere sequences to stabilize the end of a broken chromosome. We recently showed that when we remove two enzymes that normally chew away the ends of a DSB, about half of the cells acquire a new telomere at the broken end. We now would like to monitor the kinetics of telomere addition and to learn more about the proteins required for this important process.
Cell cycle regulation in response to DNA damage.
We also study the ways that a cell "knows" that there is DNA damage and how it then arrests cell growth until that damage is repaired. The control of the DNA damage "check point" is not fully understood. What is the actual signal that tells the cell it has a broken or damaged chromosome, and what is the signal that perpetuates cell cycle arrest? How do cells know when to resume growth when damage is repaired? We are analyzing mutants of yeast that fail to respond normally to these checkpoint signals.
We have been studying the phenomenon of adaptation, where cells that have an unrepaired (and unrepairable) DSB will eventually escape from the G2/M DNA damage arrest checkpoint and resume growth, despite the continued presence of the broken chromosome. We have identified a number of adaptation-defective mutations where cells remain permanently arrested. Some of these mutations are also defective in resuming cell division even after DNA damage is repaired. Some of these mutants prevent the cell from turning off the checkpoint (for example defects in two phosphatases that have to revers the phosphorylations imposed by checkpoint protein kinases). Some mutants appear to enhance end-resection and generate more signal. But we do not yet understand how many other adaptation mutants that we have found fit into this scheme. One interesting class appears to enhance the cytoplasmic degradation of normally nuclear proteins by autophagy.
Selected Publications
- Chromosome structure due to phospho-mimetic H2A modulates DDR through increased chromatin mobility. Garcia Fernandez F, Lemos B, Khalil Y, Batrin R, Haber JE, (2021) Fabre E. J. Cell Sci. 23, 258500.
- Structural parameters of palindromic repeats determine the specificity of nuclease attack of secondary structures. Saada, A, Anissia, A; Costa, A, Guo, W, Sheng, Z; Haber, JE, Lobachev, K. (2021) Nucleic Acids Res. 49, 3932-3947.
- Local nucleosome dynamics and eviction following a double-strand break are reversible by NHEJ-mediated repair in the absence of DNA replication. Tripuraneni, V , Memisoglu, G , MacAlpine, HK , Zhu, W , Tran, T , Hartemink, AJ , Haber, JE, and MacAlpine, DM (2021). Genome Research 31, 775-788. doi:10.1101/gr.271155.120.
- Loop extrusion as a mechanism for DNA damage repair foci formation. Coline Arnould, Vincent Rocher, Anne-Laure Finoux, Thomas Clouaire, Kevin Li, Felix Zhou, Pierre Caron, Philippe. E. Mangeot, Emiliano P. Ricci, Raphael Mourad, James E Haber, Daan Noordermeer, Gaëlle Legube. Nature. 2021 Feb 1; 590(7847): 660–665. Published online 2021 Feb 17. doi: 10.1038/s41586-021-03193-z
- A Rad51-independent pathway promotes single-strand template repair in gene editing.
Danielle N. Gallagher, Nhung Pham, Annie M. Tsai, Abigail N. Janto, Jihyun Choi, Grzegorz Ira, James E. Haber. PLoS Genet. 2020 Oct; 16(10): e1008689. Published online 2020 Oct 15. doi: 10.1371/journal.pgen.1008689 - Patterns of somatic structural variation in human cancer genomes. Yilong Li, Nicola D. Roberts, Jeremiah A. Wala, Ofer Shapira, Steven E. Schumacher, Kiran Kumar, Ekta Khurana, Sebastian Waszak, Jan O. Korbel, James E. Haber, Marcin Imielinski, PCAWG Structural Variation Working Group, Joachim Weischenfeldt, Rameen Beroukhim, Peter J. Campbell, PCAWG Consortium. Nature. 2020; 578(7793): 112–121. Published online 2020 Feb 5. doi: 10.1038/s41586-019-1913-9
- Mechanisms restraining break-induced replication at two-ended DNA double-strand breaks. Pham, N, Yan, Z, Yu, Y, Afreen, MF, Malkova, A, Haber, JE and Ira, G (2020) EMBO J 16, e1008689. doi: 10.1371/journal.pgen.1008689.
- Yeast ATM and ATR kinases use different mechanisms to spread histone H2A phosphorylation around a DNA double-strand break. Li K, Bronk G, Kondev J, Haber JE. (2020) PNAS, 117, 21354-21363.
- Network Rewiring of Homologous Recombination Enzymes during Mitotic Proliferation and Meiosis.
Philipp Wild, Aitor Susperregui, Ilaria Piazza, Christian Dörig, Ashwini Oke, Meret Arter, Miyuki Yamaguchi, Alexander T. Hilditch, Karla Vuina, Ki Choi Chan, Tatiana Gromova, James E. Haber, Jennifer C. Fung, Paola Picotti, Joao Matos. Mol Cell. 2019 Aug 22; 75(4): 859–874.e4. doi: 10.1016/j.molcel.2019.06.022 - Live cell monitoring of double strand breaks in S. cerevisiae. Waterman DP, Zhou F, Li K, Lee CS, Tsabar M, Eapen VV, Mazzella A, Haber JE. PLoS Genet. 2019 Mar 1;15(3):e1008001. doi: 10.1371/journal.pgen.1008001. eCollection 2019 Mar.
- PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Memisoglu G, Eapen VV, Yang Y, Klionsky DJ, Haber JE. Proc Natl Acad Sci U S A. 2019 Jan 29;116(5):1613-1620. doi: 10.1073/pnas.1817078116. Epub 2019 Jan 17.
- Mating-type switching by homology-directed recombinational repair: a matter of choice. Thon G, Maki T, Haber JE, Iwasaki H. Curr Genet. 2019 Apr;65(2):351-362. doi: 10.1007/s00294-018-0900-2.
- New insights into donor directionality of mating-type switching in Schizosaccharomyces pombe. Maki T, Ogura N, Haber JE, Iwasaki H, Thon G. PLoS Genet. 2018 May 31;14(5):e1007424. doi: 10.1371/journal.pgen.1007424. eCollection 2018 May.
- Multiplexed precision genome editing with trackable genomic barcodes in yeast. Roy KR, Smith JD, Vonesch SC, Lin G, Tu CS, Lederer AR, Chu A, Suresh S, Nguyen M, Horecka J, Tripathi A, Burnett WT, Morgan MA, Schulz J, Orsley KM, Wei W, Aiyar RS, Davis RW, Bankaitis VA, Haber JE, Salit ML, St Onge RP, Steinmetz LM. Nat Biotechnol. 2018 Jul;36(6):512-520. doi: 10.1038/nbt.4137. Epub 2018 May 7.
- DNA Repair: The Search for Homology. Haber JE. Bioessays. 2018 May;40(5):e1700229. doi: 10.1002/bies.201700229.
- Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Anand R, Beach A, Li K and Haber J (2017). Nature. 544, 377–380.
- A pathway of targeted autophagy is induced by DNA damage in budding yeast. Eapen VV, Waterman DP, Bernard A, Schiffmann N, Sayas E, Kamber R, Lemos B, Memisoglu G, Ang J, Mazella A, Chuartzman SG, Loewith RJ, Schuldiner M, Denic V, Klionsky DJ and Haber JE (2017). Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):E1158-E1167.
- Homology Requirements and Competition between Gene Conversion and Break-Induced Replication during Double-Strand Break Repair. Mehta A, Beach A and Haber JE (2017). Mol Cell 65(3): 515-526 e513.
- Chromosome-refolding model of mating-type switching in yeast. Avsaroglu B, Bronk G, Li K, Haber JE and Kondev J (2016). Proc Natl Acad Sci USA: 201607103.
- A Life Investigating Pathways That Repair Broken Chromosomes. Haber JE. (2016). Annu Rev Genet. 50:1-28.
- The rule of three. Haber JE. (2016). Nat Rev Mol Cell Biol 17(6): 333.
- Sgs1 and Mph1 Helicases Enforce the Recombination Execution Checkpoint During DNA Double-Strand Break Repair in Saccharomyces cerevisiae. Jain S, Sugawara N, Mehta A, Ryu T and Haber JE (2016). Genetics 203(2): 667-675.
- The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. Jasin M and Haber JE (2016). DNA Repair (Amst) 44:6-16.
- Re-establishment of nucleosome occupancy during double-strand break repair in budding yeast. Tsabar M, Hicks WM, Tsaponina O and Haber JE (2016). DNA Repair (Amst). 47:21-29
- Asf1 facilitates dephosphorylation of Rad53 after DNA double-strand break repair. Tsabar, M., D. P. Waterman, F. Aguilar, L. Katsnelson, V. V. Eapen, G. Memisoglu and J. E. Haber (2016). Genes Dev 30(10): 1211-1224.
- MTE1 Functions with MPH1 in Double-Strand Break Repair. Yimit, A., T. Kim, R. P. Anand, S. Meister, J. Ou, J. E. Haber, Z. Zhang and G. W. Brown (2016). Genetics 203(1): 147-157.
- Role of Double-Strand Break End-Tethering during Gene Conversion in Saccharomyces cerevisiae. Jain S, Sugawara, N and Haber, J. (2016). Plos Genetics 12(4): e1005976.
- Chromosome position determines the success of double-strand break repair. Lee CS, Wang RW, Chang HH, Capurso D, Segal MR, Haber JE. (2016). Proc Natl Acad Sci U S A 113(2): E146-154.
- Chromosomes at loose ends. Nakajima Y, Haber JE. (2016). Nat Cell Biol 18(3): 257-259.
- Sgs1 and Mph1 Helicases Enforce the Recombination Execution Checkpoint During DNA Double-Strand Break Repair in Saccharomyces cerevisiae. Jain S, Sugawara N, Mehta A, Ryu T, Haber JE. (2016). Genetics. June 1, 2016 vol. 203 no. 2 667-675; 115.184317 2016 Apr 13.
- A Cohesin-Based Partitioning Mechanism Revealed upon Transcriptional Inactivation of Centromere. Tsabar M, Haase J, Harrison B, Snider CE, Eldridge B, Kaminsky L, Hine RM, Haber JE, Bloom K. (2016). Plos Genetics 12(4): e1006021.
- Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Tsabar M, Eapen VV, Mason JM, Memisoglu G, Waterman DP, Long MJ, Bishop DK, Haber JE. (2015). Nucleic Acids Res 43(14): 6889-6901.
- Caffeine inhibits gene conversion by displacing Rad51 from ssDNA. Tsabar M, Mason JM, Chan YL, Bishop DK, Haber JE. (2015). Nucleic Acids Res 43(14): 6902-6918.
- Break-induced replication repair of damaged forks induces genomic duplications in human cells. Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD. (2014). Science 343(6166): 88-91.
- Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Lee CS, Lee K, Legube G, Haber JE. (2014). Nat Struct Mol Biol 21(1): 103-109.
- Sources of DNA double-strand breaks and models of recombinational DNA repair. Mehta A, Haber JE. (2014). Cold Spring Harb Perspect Biol 6(9): a016428.
- Frequent Interchromosomal Template Switches during Gene Conversion in S. cerevisiae. Tsaponina O, Haber JE. (2014). Mol Cell 55(4): 615-625.
- Chromosome rearrangements via template switching between diverged repeated sequences. Anand RP, Tsaponina O, Greenwell PW, Lee CS, Du W, Petes TD, Haber JE. (2014). Genes Dev 28(21): 2394-2406.
- Effect of chromosome tethering on nuclear organization in yeast. Avşaroğlu B, Bronk G, Gordon-Messer S, Ham J, Bressan DA, Haber JE, Kondev J. (2014). PLoS ONE 9(7): e102474.
- DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase. Dotiwala F, Eapen VV, Harrison JC, Arbel-Eden A, Ranade V, Yoshida S, Haber JE. (2013). Proc Natl Acad Sci USA 110(1): E41-49.
- DNA damage signaling triggers the cytoplasm-to-vacuole pathway of autophagy to regulate cell cycle progression. Eapen VV, Haber JE. (2013). Autophagy 9(3): 440-441.
- Systematic triple-mutant analysis uncovers functional connectivity between pathways involved in chromosome regulation. Haber JE, Braberg H, Wu Q, Alexander R, Haase J, Ryan C, Lipkin-Moore Z, Franks-Skiba KE, Johnson T, Shales M, Lenstra TL, Holstege FC, Johnson JR, Bloom K, Krogan NJ. (2013). Cell Rep 3(6): 2168-2178.
- Migrating bubble during break-induced replication drives conservative DNA synthesis." Nature 502(7471): 389-392.
- Break-induced DNA replication. Anand RP, Lovett ST, Haber JE. (2013). Cold Spring Harb Perspect Biol 5(12): a010397.
- The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Eapen VV, Sugawara N, Tsabar M, Wu WH, Haber JE. (2012). Mol Cell Biol 32(22): 4727-4740.
- Mutations arising during repair of chromosome breaks. Malkova A, Haber JE. (2012). Annu Rev Genet 46: 455-473.
- Mating-type genes and MAT switching in Saccharomyces cerevisiae. Haber JE. Genetics. 2012 May;191(1):33-64.
- Regulation of budding yeast mating-type switching donor preference by the FHA domain of Fkh1. Li J, Coïc E, Lee K, Lee CS, Kim JA, Wu Q, Haber JE. PLoS Genet. 2012 Apr;8(4):e1002630.
- Dynamics of homology searching during gene conversion in Saccharomyces cerevisiae revealed by donor competition. Coïc E, Martin J, Ryu T, Tay SY, Kondev J, Haber JE. Genetics. 2011 Dec;189(4):1225-33.
- QnAs with James E. Haber. Haber JE. Proc Natl Acad Sci U S A. 2011 Apr 5;108(14):5479.
- Real-time analysis of double-strand DNA break repair by homologous recombination. Hicks WM, Yamaguchi M, Haber JE. Proc Natl Acad Sci U S A. 2011 Feb 22;108(8):3108-15.
- Protein phosphatases pph3, ptc2, and ptc3 play redundant roles in DNA double-strand break repair by homologous recombination. Kim JA, Hicks WM, Li J, Tay SY, Haber JE. Mol Cell Biol. 2011 Feb;31(3):507-16.
- Fast live simultaneous multiwavelength four-dimensional optical microscopy. Carlton PM, Boulanger J, Kervrann C, Sibarita JB, Salamero J, Gordon-Messer S, Bressan D, Haber JE, Haase S, Shao L, Winoto L, Matsuda A, Kner P, Uzawa S, Gustafsson M, Kam Z, Agard DA, Sedat JW. Proc Natl Acad Sci U S A. 2010 Sep 14;107(37):16016-22.
- Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Hicks WM, Kim M, Haber JE. Science. 2010 Jul 2;329(5987):82-5.
- Sgs1 and exo1 redundantly inhibit break-induced replication and De Novo telomere addition at broken chromosome ends. Lydeard JR, Lipkin-Moore Z, Jain S, Eapen VV, Haber JE. PLoS Genet. 2010 May 27;6(5):e1000973.
- Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Lydeard JR, Lipkin-Moore Z, Sheu YJ, Stillman B, Burgers PM, Haber JE. Genes Dev. 2010 Jun 1;24(11):1133-44.
- Mec1/Tel1-dependent phosphorylation of Slx4 stimulates Rad1-Rad10-dependent cleavage of non-homologous DNA tails. Toh GW, Sugawara N, Dong J, Toth R, Lee SE, Haber JE, Rouse J. DNA Repair (Amst). 2010 Jun 4;9(6):718-26.
- Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. Saponaro M, Callahan D, Zheng X, Krejci L, Haber JE, Klein HL, Liberi G. PLoS Genet. 2010 Feb 26;6(2):e1000858.
- Mad2 prolongs DNA damage checkpoint arrest caused by a double-strand break via a centromere-dependent mechanism. Dotiwala F, Harrison JC, Jain S, Sugawara N, Haber JE. Curr Biol. 2010 Feb 23;20(4):328-32.
- Replicon dynamics, dormant origin firing, and terminal fork integrity after double-strand break formation. Doksani Y, Bermejo R, Fiorani S, Haber JE, Foiani M. Cell. 2009 Apr 17;137(2):247-58.
- A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. Genes Dev. 2009 Feb 1;23(3):291-303.
- Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Kim JA, Haber JE. Proc Natl Acad Sci U S A. 2009 Jan 27;106(4):1151-6.
- Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, et al. Genes Dev. 2009;23(1):67-79.
- Chromatin assembly factors Asf1 and CAF-1 have overlapping roles in deactivating the DNA damage checkpoint when DNA repair is complete. Kim JA, Haber JE. Proc Natl Acad Sci U S A. 2009;106(4):1151-6.
- Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, et al. EMBO J. 2008;27(10):1502-12.
- Functional interactions between Sae2 and the Mre11 complex. Kim HS, Vijayakumar S, Reger M, Harrison JC, Haber JE, Weil C, et al. Genetics. 2008;178(2):711-23.
- Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. EMBO J. 2008;27(14):1953-62.
- Alternative endings. Haber JE. Proc Natl Acad Sci U S A. 2008;105(2):405-6.
- Mechanisms of Rad52-independent spontaneous and UV-induced mitotic recombination in Saccharomyces cerevisiae. Coic E, Feldman T, Landman AS, Haber JE. Genetics. 2008;179(1):199-211.
- Anaphase onset before complete DNA replication with intact checkpoint responses. Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A, Machin F, et al. Science. 2007; 315 (5817): 1411-5.
- Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Morrison AJ, Kim JA, Person MD, Highland J, Xiao J, Wehr TS, et al. Cell. 2007;130(3):499-511.
- Break-induced replication and telomerase-independent telomere maintenance require Pol32. Lydeard JR, Jain S, Yamaguchi M, Haber JE. Nature. 2007;448(7155):820-3.
- Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. J Cell Biol. 2007;178(2):209-18.
- Phosphorylation of Slx4 by Mec1 and Tel1 regulates the single-strand annealing mode of DNA repair in budding yeast. Flott S, Alabert C, Toh GW, Toth R, Sugawara N, Campbell DG, et al. Mol Cell Biol. 2007;27(18):6433-45.
- The yeast DNA damage checkpoint proteins control a cytoplasmic response to DNA damage. Dotiwala F, Haase J, Arbel-Eden A, Bloom K, Haber JE. Proc Natl Acad Sci U S A. 2007;104(27):11358-63.
- SMC proteins, new players in the maintenance of genomic stability. Cortes-Ledesma F, de Piccoli G, Haber JE, Aragon L, Aguilera A. Cell Cycle. 2007;6(8):914-8.