Yerbol Z. Kurmangaliyev
 Assistant Professor of Biology
Assistant Professor of Biology
Research Description
How are brain connectomes encoded in the genome?
Brain function relies on proper wiring of neural circuits. There are a myriad of neurons (from 100,000 in fruit flies to 100 billion in humans) that can recognize and form connections with specific partners in the immensely crowded environment of developing brains. The precise wiring diagrams of neural circuits are thought to be directly encoded by genetic programs of brain development. Errors in these programs can result in miswired circuits leading to brain disorders.
How are complex brains built during development? What are the genetic and molecular mechanisms behind the precise wiring of neural circuits that underlie the complex cognition and behaviors of animals? The developmental and evolutionary origin of brain complexity remains one of the great mysteries in biology. We aim to address these questions from a genomic perspective. We are interested in understanding how genes and genomes encode the architecture of neural circuits.
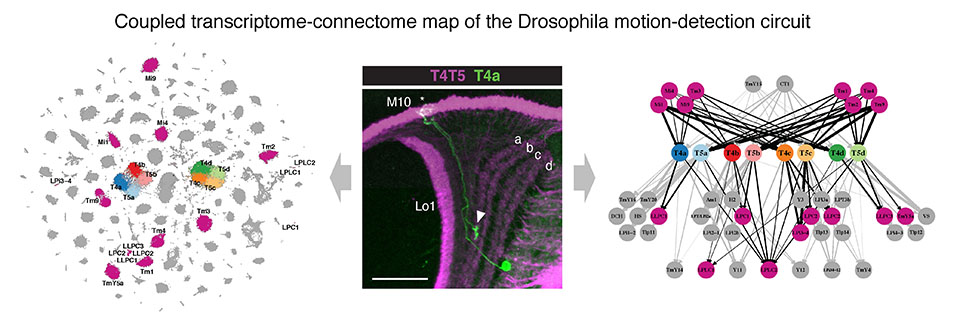
Our main model system is the Drosophila melanogaster brain. It offers nearly complete connectivity maps and genetic access to hundreds of neuronal cell types. Our lab combines the power of genomic technologies with the rich genetic toolbox of fruit flies to study molecular mechanisms underlying brain development. Today, advances in single-cell genomics and brain connectomics allow the generation of coupled transcriptome-connectome maps of the developing circuits. They provide a nearly complete description of transcriptomes for both sides of every synaptic connection. We build and use these maps to study how neural circuits emerge during brain development and evolution. Our approaches include both genome-wide computational studies and hypothesis-driven in vivo studies. Ultimately, cracking the molecular code and logic of brain wiring could enable us to rewire neural circuits and behaviors in predictable and interpretable ways.
Selected Publications
- Yoo, J., Dombrovski, M., Mirshahidi, P., Nern, A., LoCascio, S. A., Zipursky, S. L., & Kurmangaliyev, Y. Z. (2023). Brain wiring determinants uncovered by integrating connectomes and transcriptomes. Current biology, 33:3998–4005.e6.
- Dombrovski, M., Peek, M. Y., Park, J. Y., Vaccari, A., Sumathipala, M., Morrow, C., Breads, P., Zhao, A., Kurmangaliyev, Y. Z., Sanfilippo, P., Rehan, A., Polsky, J., Alghailani, S., Tenshaw, E., Namiki, S., Zipursky, S. L., & Card, G. M. (2023). Synaptic gradients transform object location to action. Nature, 613:534–542.
- McCourt, J. L., Stearns-Reider, K. M., Mamsa, H., Kannan, P., Afsharinia, M. H., Shu, C., Gibbs, E.M., Shin, K. M., Kurmangaliyev, Y. Z., Schmitt, L. R., Hansen, K. C., & Crosbie, R. H. (2023). Multi-omics analysis of sarcospan overexpression in mdx skeletal muscle reveals compensatory remodeling of cytoskeleton-matrix interactions that promote mechanotransduction pathways. Skeletal muscle, 13:1.
- Stearns-Reider, K. M., Hicks, M. R., Hammond, K. G., Reynolds, J. C., Maity, A., Kurmangaliyev, Y. Z., Chin, J., Stieg, A. Z., Geisse, N. A., Hohlbauch, S., Kaemmer, S., Schmitt, L. R., Pham, T. T., Yamauchi, K., Novitch, B. G., Wollman, R., Hansen, K. C., Pyle, A. D., & Crosbie, R. H. (2023). Myoscaffolds reveal laminin scarring is detrimental for stem cell function while sarcospan induces compensatory fibrosis. NPJ Regenerative medicine, 8:16.
- Jain, S.*, Lin, Y.*, Kurmangaliyev, Y. Z., Valdes-Aleman J., LoCascio, S. A., Mirshahidi, P., Parrington, B., & Zipursky, S. L. (2022) A global timing mechanism regulates cell-type-specific wiring programmes. Nature, 604:112-118.
- Kurmangaliyev, Y. Z., Yoo, J., Valdes-Aleman, J., Sanfilippo, P., & Zipursky, S. L. (2020) Transcriptional Programs of Circuit Assembly in the Drosophila Visual System. Neuron, 108:1–13.
- Yokota, T., McCourt, J., Ma, F., Ren, S., Li, S., Kim, T. H., Kurmangaliyev, Y. Z., Nasiri, R., Ahadian, S., Nguyen, T., Tan, X., Zhou, Y., Wu, R., Rodriguez, A., Cohn, W., Wang, Y., Whitelegge, J., Ryazantsev, S., Khademhosseini, A., Teitell, M. A., Chiou, P.-Y., Birk, D. E., Rowat, A. C., Crosbie, R.-H., Pellegrini, M., Seldin, M., Lusis, A. J., & Deb, A. (2020). Type V Collagen in Scar Tissue Regulates the Size of Scar after Heart Injury. Cell, 182:545–562.
- Kurmangaliyev, Y. Z.*, Yoo, J.*, LoCascio, S. A.*, & Zipursky, S. L. (2019). Modular transcriptional programs separately define axon and dendrite connectivity. eLife, 8, e50822.
- Romero, Z., Lomova, A., Said, S., Miggelbrink, A., Kuo, C. Y., Campo-Fernandez, B., Hoban, M. D., Masiuk, K. E., Clark, D. N., Long, J., Sanchez, J. M., Velez, M., Miyahira, E., Zhang, R., Brown, D., Wang, X., Kurmangaliyev, Y. Z., Hollis, R. P., & Kohn, D. B. (2019). Editing the Sickle Cell Disease Mutation in Human Hematopoietic Stem Cells: Comparison of Endonucleases and Homologous Donor Templates. Molecular therapy, 27:1389–1406.
- Sarin, S.*, Zuniga-Sanchez, E.*, Kurmangaliyev, Y. Z., Cousins, H., Patel, M., Hernandez, J., Zhang, K. X., Samuel, M. A., Morey, M., Sanes, J. R., & Zipursky, S. L. (2018) Role for Wnt Signaling in Retinal Neuropil Development: Analysis via RNA-Seq and In Vivo Somatic CRISPR. Mutagenesis. Neuron, 98:109–126.
- Kurmangaliyev, Y. Z., Ali, S., & Nuzhdin S. V. (2016) Genetic Determinants of RNA Editing Levels of ADAR Targets in Drosophila melanogaster. G3 (Bethesda), 6:391–396.
- Kurmangaliyev, Y. Z., Favorov, A.V., Osman, N. M., Lehmann, K. V., Campo, D., Salomon, M. P., Tower, J., Gelfand, M. S., & Nuzhdin, S. V. (2015) Natural variation of gene models in Drosophila melanogaster. BMC Genomics, 16:198.
- Kurmangaliyev, Y. Z., Sutormin, R. A., Naumenko, S. A., Bazykin, G. A., & Gelfand, M. S. (2013) Functional implications of splicing polymorphisms in the human genome. Human Molecular Genetics, 22:3449–3459.
- Kurmangaliyev, Y. Z., Goland, A., Gelfand, M. S. (2011) Evolutionary patterns of phosphorylated serines. Biology Direct, 6:8.
- Kurmangaliyev, Y. Z. & Gelfand, M. S. (2008) Computational analysis of splicing errors and mutations in human transcripts. BMC Genomics, 9:13.
- Deev, I. E., Vasilenko, K. P., Kurmangaliev, E. Zh., Serova, O. V., Popova, N. V., Galagan, Y. S., Burova, E. B., Zozulya, S. A., Nikol'skii, N. N., Petrenko, A. G. (2006) Effect of changes in ambient pH on phosphorylation of cellular proteins. Dokl Biochem Biophys (Moscow), 408:184–187.