Michael Marr
 Professor of Biology
Professor of Biology
Research Description
Mechanisms Controlling Gene Expression
My lab is focused on investigating how a metazoan cell responds to developmental and environmental signals and how this response is manifest in changes in gene expression. The developmental, metabolic, and environmental signals a cell receives induce changes in both RNA and protein synthesis. Strict control of these pathways is required for integrity of homeostasis at both the cellular and the organismal level. Loss of this control leads to disease and developmental abnormalities. As a result, organisms and cells control these pathways at both the transcriptional and post transcriptional level. I utilize two model systems to investigate both levels of regulation of gene expression. For the investigation of a robust transcriptional response, I utilize the Drosophila metallothionein genes and the cellular response to heavy metal shock. For experiments investigating post-transcriptional regulation, I use the Drosophila insulin-like receptor pathway and the cellular response to insulin.
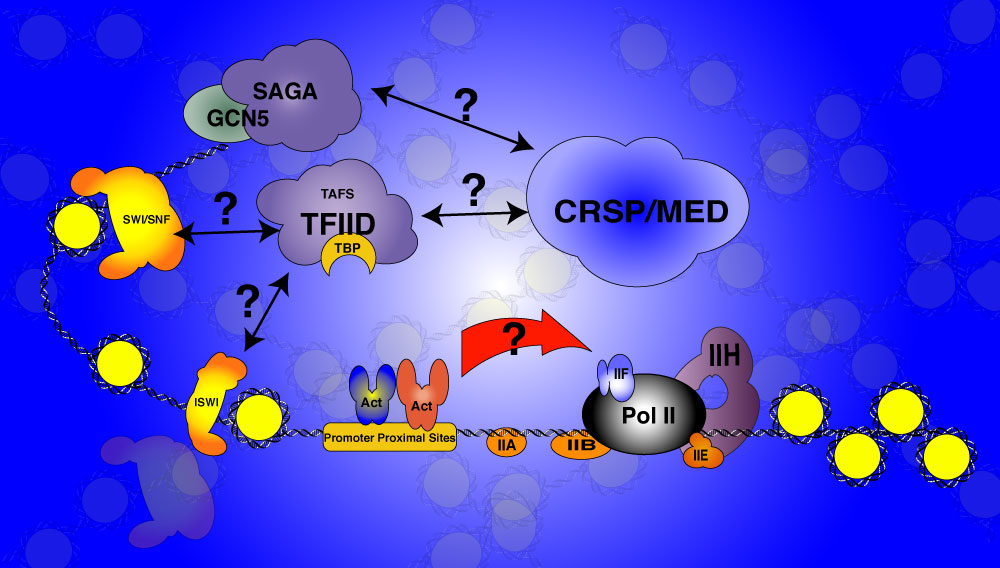
Transcriptional control of gene expression
The cellular response to heavy metal shock is mediated directly by a metal dependent transcription factor (MTF-1) and is primarily transcriptional. This provides an excellent model system for investigating the mechanisms governing transcriptional responses to cellular signals. I use a combination of RNA interference (RNAi), biochemical assays, and single cell immunofluorescence to investigate the role of many of the cofactors required for transcription activation. Some of the questions we are trying to address are: What steps are controlled in vivo by the various co-factors? Are the roles redundant? How many pathways are there to activation of transcription?
Post-transcriptional control of gene expression
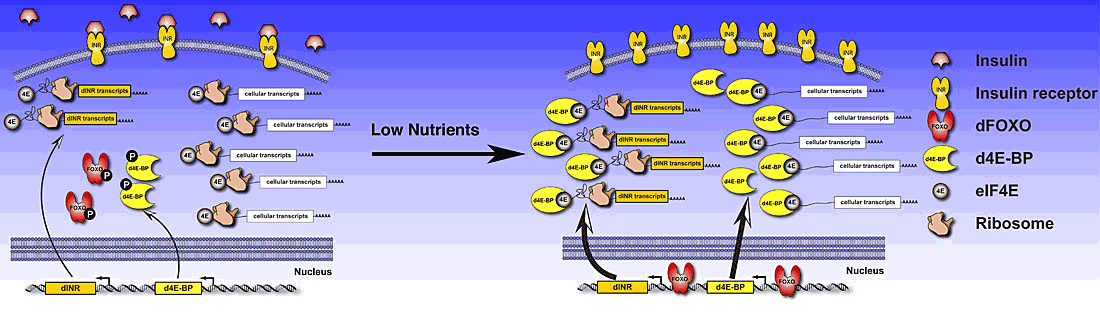
The insulin receptor pathway is a classical signal transduction pathway that plays important roles in governing cell and organ size in metazoans and also metabolic homeostasis and even lifespan. Interestingly, the gene for insulin receptor (dINR) is a direct transcriptional target of the pathway. A second important target gene is the global translation inhibitor eIF4E-binding protein (4E-BP). This discovery highlights one mechanism of insulin-mediated control of protein synthesis. This finding also exposes an apparent inconsistency. Under these conditions 4E-BP is repressing cap-dependent translation but the dINR protein continues to be synthesized. These findings were reconciled once we found that the dINR utilizes an internal ribosome entry site (IRES) that allows cap-independent translation initiation. This finding demonstrates a novel functional coupling of transcription and translation that allows amplification of the insulin receptor pathway specific target genes. The pathway activates 4E-BP and at the same time activates the transcription of genes that are resistant to 4E-BP. This leads to an effective switch of the cellular machinery to translation of targets of the pathway. Currently we are trying to understand how this cellular IRES engages the translational machinery. In addition we are trying to determine how often this type of functional coupling is utilized.
Some of the questions our lab is trying to answer. Do the co-activators communicate with one another? Are they redundant? What does it mean for an activator to communicate with the basal machinery?
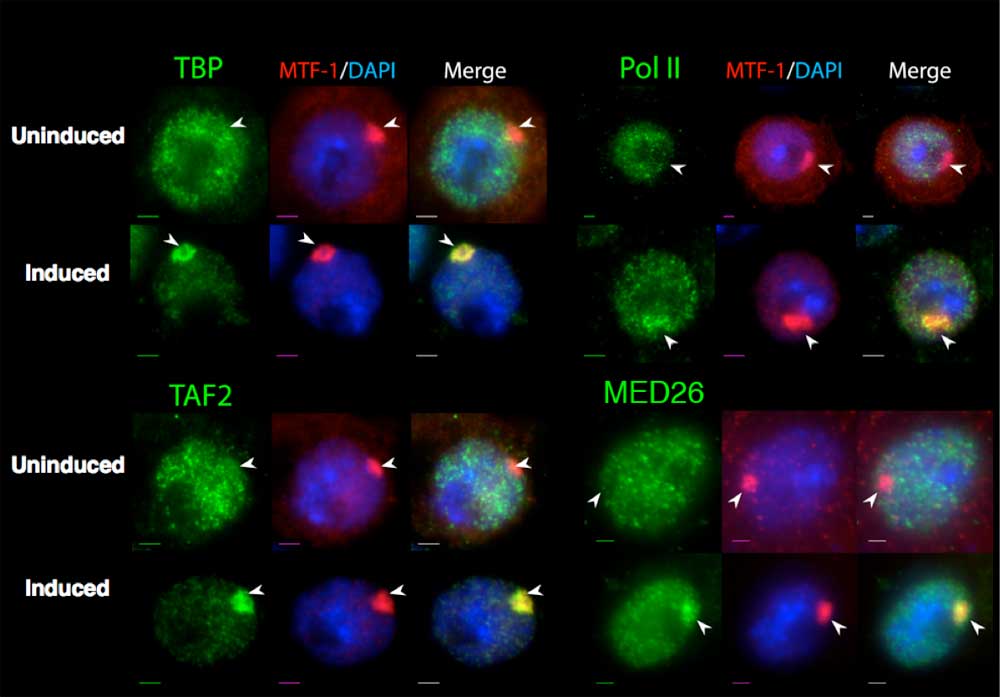
Recruitment of coactivators to the transgenic locus. In situ staining to determine protein recruitment to the transgenic MtnA cluster. The protein specified above the first column in each panel was detected with an FITC conjugated secondary antibody. In the middle column of each panel, MTF-1 was detected with a Rhodamin-X conjugated secondary antibody, and DNA was stained with DAPI. The third column shows the merge of these channels. In each panel, the top row shows a cell in the absence of copper; the bottom row shows a cell in the presence of copper for 2 or 4 hr. Scale bar, 2 µm.
Model of the role of IRES activity in production of Drosophila insulin receptor (dINR). On the right, in conditions of high nutrients or insulin-like peptides, dFOXO and d4E-BP are phosphorylated and inactive. This results in a basal level of expression of the dINR and 4E-BP genes. The translational machinery is divided evenly between dINR and other cellular transcripts. On the left, in conditions of low nutrients or insulin-like peptides, dFOXO and d4E-BP are unphosphorylated. dFOXO can activate transcription of both the dINR and d4E-BP genes. The unphosphorylated d4E-BP is able to bind to 4E, preventing the translation of cellular cap-dependent translation. This results in an increase in the dINR transcript and an uneven distribution of the translation machinery amplifying expression of dINR at both the transcription and translation levels. This results in the accumulation of dINR in the cellular membrane, poising the cell for a rapid response to signals when nutrients are again available.
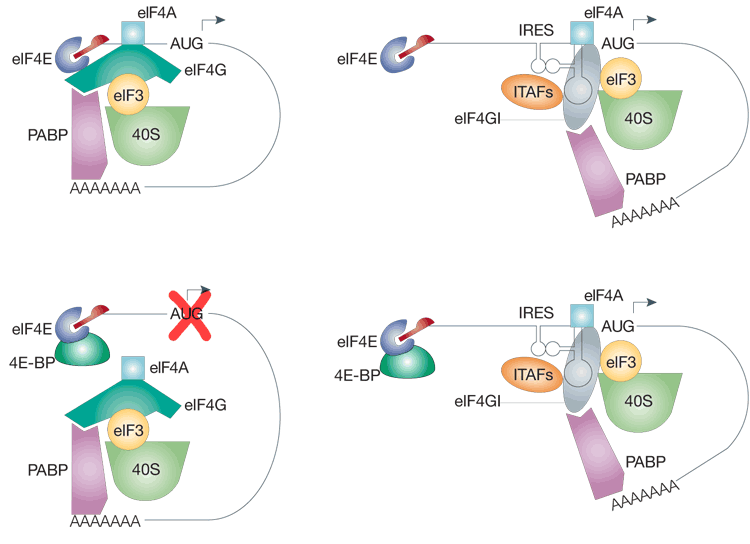
Models for translation initiation
Under normal growth conditions both cap-dependent and cap-independent (IRES) mechanisms of translation initiation are utilized (top panel). In stress conditions such as low nutrients 4E-BP prevents cap-dependent initiation but transcripts containing an IRES are still actively translated (bottom panel). [Modified from Holcik & Sonenberg, Nat. Rev. Mol. Cell. Biol. 2005]
Selected Publications
- TFIID Enables RNA Polymerase II Promoter-Proximal Pausing. Fant CB, Levandowski CB, Gupta K, Maas ZL, Moir J, Rubin JD, Sawyer A, Esbin MN, Rimel JK, Luyties O, Marr MT, Berger I, Dowell RD, Taatjes DJ. Mol Cell. 2020 May 21;78(4):785-793.e8. doi: 10.1016/j.molcel.2020.03.008.
- Complete Genome Sequences of Two Vibrio natriegens Bacteriophages. Harris MT, Ho TC, Fruchtman H, Garin ME, Kubatin V, Lu T, Xue L, Marr MT 2nd. Microbiol Resour Announc. 2020 Nov 5;9(45):e01133-20. doi: 10.1128/MRA.01133-20.
- The Metastasis Suppressor Protein Nme1 Is a Concentration-Dependent Modulator of Ca2+/Calmodulin-Dependent Protein Kinase II. Royer L, Shangraw K, Herzog JJ, Pouvreau S, Marr MT 2nd, Paradis S. Biochemistry. 2019 Jun 18;58(24):2710-2714. doi: 10.1021/acs.biochem.9b00121.
- The Ras-like GTPase Rem2 is a potent inhibitor of calcium/calmodulin-dependent kinase II activity. Royer L, Herzog JJ, Kenny K, Tzvetkova B, Cochrane JC, Marr MT 2nd, Paradis S. J Biol Chem. 2018 Sep 21;293(38):14798-14811. doi: 10.1074/jbc.RA118.003560.
- The GCN2-ATF4 Signaling Pathway Induces 4E-BP to Bias Translation and Boost Antimicrobial Peptide Synthesis in Response to Bacterial Infection. Vasudevan D, Clark NK, Sam J, Cotham VC, Ueberheide B, Marr MT 2nd, Ryoo HD. Cell Rep. 2017 Nov 21;21(8):2039-2047.
- Rem2 signaling affects neuronal structure and function in part by regulation of gene expression. Kenny K, Royer L, Moore AR, Chen X, Marr MT 2nd, Paradis S. Mol Cell Neurosci. 2017 Dec;85:190-201.
- 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. Kang MJ, Vasudevan D, Kang K, Kim K, Park JE, Zhang N, Zeng X, Neubert TA, Marr MT 2nd, Ryoo HD. J Cell Biol. 2017 Jan 2;216(1):115-129.
- dFOXO Activates Large and Small Heat Shock Protein Genes in Response to Oxidative Stress to Maintain Proteostasis in Drosophila. Donovan MR, Marr MT 2nd. J Biol Chem. 2016 Sep 2;291(36):19042-50.
- Ubiquilin-mediated Small Molecule Inhibition of Mammalian Target of Rapamycin Complex 1 (mTORC1) Signaling. Coffey RT, Shi Y, Long MJ, Marr MT 2nd, Hedstrom L (2016). J Biol Chem 291(10): 5221-5233.
- FOXO regulates RNA interference in Drosophila and protects from RNA virus infection. Spellberg MJ and MT Marr 2nd (2015). Proc Natl Acad Sci U S A. 2015 Nov 24;112(47):14587-92.
- "The metazoan-specific mediator subunit 26 (Med26) is essential for viability and is found at both active genes and pericentric heterochromatin in Drosophila melanogaster. Marr SK, Lis JT, Treisman JE, Marr MT 2nd (2014). Mol Cell Biol 34(14): 2710-2720.
- CaMKII-dependent phosphorylation of the GTPase Rem2 is required to restrict dendritic complexity. Ghiretti AE, Kenny K, Marr MT 2nd, Paradis S (2013). J Neurosci 33(15): 6504-6515.
- The insulin receptor cellular IRES confers resistance to eIF4A inhibition. Olson CM, Donovan MR, Spellberg MJ, Marr MT 2nd (2013). Elife. 2013 Jul 16;2:e00542.
- Holo-TFIID controls the magnitude of a transcription burst and fine-tuning of transcription. Pennington KL, Marr SK, Chirn GW, Marr MT 2nd (2013). Proc Natl Acad Sci U S A 110(19): 7678-7683.
- Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, Rosbash M. (2011). Genes Dev 25(23): 2502-2512.
- Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. Kadener S, Rodriguez J, Abruzzi KC, Khodor YL, Sugino K, Marr MT 2nd, Nelson S, Rosbash M (2009). RNA. 2009 Apr;15(4):537-45.
- TAF4 takes flight. Marr MT 2nd. (2009) Proc Natl Acad Sci U S A. 2009 Feb 3;106(5):1295-6.
- MyoD targets TAF3/TRF3 to activate myogenin transcription. Deato MD, Marr MT, Sottero T, Inouye C, Hu P, Tjian R. (2008) Mol Cell. 2008 Oct 10;32(1):96-105.
- TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Hsu JY, Juven-Gershon T, Marr MT 2nd, Wright KJ, Tjian R, Kadonaga JT. Genes Dev. 2008 Sep 1;22(17):2353-8.
- The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. (2008) Nature. 2008 Feb 14;451(7180):783-8.
- Characterization of the Drosophila insulin receptor promoter. Casas-Tinto S, Marr MT 2nd, Andreu P, Puig O. (2007). Biochim Biophys Acta. 1769, 236-43.
- IRES-Mediated Coupling of Transcription and Translation Amplifies Insulin Receptor Feedback. Marr MT, D’Alessio JA, Puig O and Tjian R. (2007). Genes Dev. 21, 175-83.
- Coactivator cross-talk specifies transcriptional output. Marr MT, Isogai Y, Wright KJ and Tjian R. (2006). Genes Dev. 20, 1458-69.
- TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Wright KJ, Marr MT and Tjian R. (2006). PNAS. 103, 12347-52.
- Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. (2005) Mol Cell. 17, 103-12.
- Regulatory diversity among metazoan co-activator complexes. Taatjes DJ, Marr MT, Tjian R. (2004) Nat Rev Mol Cell Biol. 5, 403-10.
- Interactions among CII protein, RNA polymerase and the lambda PRE promoter: contacts between RNA polymerase and the –35 region of PRE are identical in the presence and absence of CII protein. Marr MT, Roberts JW, Brown SE, Klee M, Gussin GN. (2004). Nucleic Acids Res. 32, 1083-90.
- Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Puig O, Marr MT, Ruhf ML, Tjian R. (2003). Genes Dev. 17, 2006-20.
- E. coli transcription repair coupling factor (Mfd protein) rescues arrested complexes by promoting forward translocation. Park JS, Marr MT, Roberts JW. (2002). Cell. 109, 757-67.
- Restructuring of an RNA polymerase holoenzyme elongation complex by lambdoid phage Q proteins. Marr MT, Datwyler S, Meares C, Roberts JW (2001). Proc Natl Acad Sci. 98, 8972-8.
- Function of Transcription Cleavage Factors GreA and GreB at a Regulatory Pause Site. Marr MT, Roberts JW. (2000). Mol Cell. 6, 1275-85.
- The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Sharp MM, Chan CL, Lu CZ, Marr MT, Nechaev S, Merritt EW, Severinov K, Roberts JW, and Gross CA. (1999). Genes Dev. 13, 3015-26.
- Antitermination by bacteriophage lambda Q protein. Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, and Roberts CW. (1998). Cold Spring Harb Symp Quant Biol. 63, 319-25.
- A surface of Escherichia coli sigma 70 required for promoter function and antitermination by phage lambda Q protein. Ko DC, Marr MT, Guo J, and Roberts JW. (1998). Genes Dev. 12, 3276-85.
- Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Marr MT, Roberts JW. (1997). Science. 276, 1258-60.
- Pribnow Box Recognition and Melting by Escherichia coli RNA Polymerase. Darst SA, Roberts JW, Malhotra A, Marr M, Severinov K & Severinova E. (1997). In Nucleic Acids & Mol. Biol., F.E.D.M.J. Lilley, ed. (London: Springer), pp. 551-56.
- Domain organization of the Escherichia coli RNA polymerase sigma 70 subunit. Severinova E, Severinov K, Fenyo D, Marr M, Brody EN, Roberts JW, Chait BT, Darst SA. (1996). J Mol Biol. 263, 637-47.