Kaushik Ragunathan
 Assistant Professor of Biology
Assistant Professor of Biology
Research Description
Molecular mechanisms of epigenetic inheritance; single molecule approaches to study chromatin associated factors in vitro and in cells
Nothing about development in multicellular organisms makes sense except in the light of epigenetics. It captures the remarkable capacity for cells with identical genomes, such as the billion or so cells in our bodies, to differentially regulate their genes and retain these patterns of expression throughout the life of the organism. This process of establishing epigenetic gene expression states is intricately tied to how the genome is organized and packaged by proteins called histones. Molecular labels in the form of histone modifications constitute a major pathway that bookmarks gene expression states in eukaryotic cells without alterations to the underlying DNA sequence.
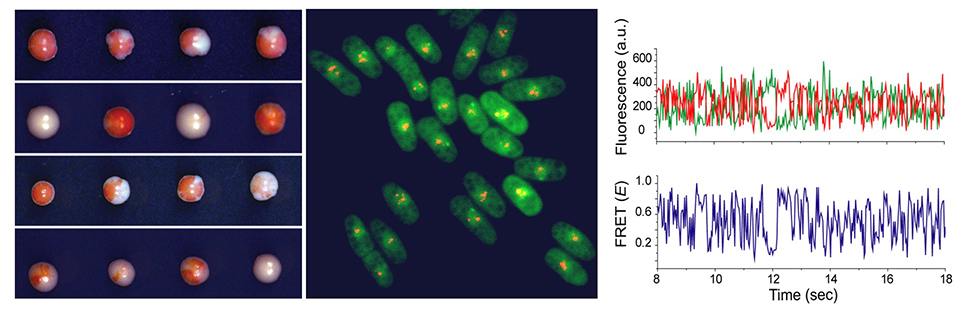
Histone H3 lysine 9 methylation (H3K9me) is associated with transcription silencing and heterochromatin formation. Fission yeast (S. pombe) has a minimalist heterochromatin architecture that is amenable to high-throughput genetics and biochemistry. A trio of conserved proteins regulates heterochromatin, which includes, 1) an H3K9me specific ''writer,'' Clr4Suv39h that catalyzes H3K9me 2) an H3K9me specific ''reader,'' Swi6HP1 that binds to H3K9me chromatin and silences transcription and, 3) an H3K9me specific ''eraser,'' Epe1JmjC, that opposes heterochromatin assembly and epigenetic inheritance. Fusing Clr4 to the tetracycline-inducible TetR DNA binding domain facilitates rapid and reversible control of heterochromatin assembly. Our innovative genetic strategy has enabled us to identify chromatin-associated factors with unique roles in heterochromatin maintenance.
We use a combination of genetic screens in yeast, biochemistry to understand the functions of specific mutants that we isolate through our screens and functional analysis of their dynamics using in vitro and in vivo single molecule approaches. We also employ whole genome based methods such as RNA-seq and ChIP-seq which helps us capture the molecular states of cells that drives changes in gene expression during homeostasis and adaptaton.
Selected Publications
2022
- Saikat Biswas, Joshua D. Karslake, Ziyuan Chen, Ali Farhat, Amanda Ames, Gulzhan Raiymbek, Peter L. Freddolino*, Julie S. Biteen*, and Kaushik Ragunathan*, “HP1 oligomerization compensates for low-affinity H3K9me recognition and provides a tunable mechanism for heterochromatin-specific localization” in revision (Science Advances)
- Kehan Bao, Chun-Min Shan, Xiao Chen, Gulzhan Raiymbek, Jeremy G. Monroe, Yimeng Fang, Takenori Toda, Kristin S. Koutmou, Kaushik Ragunathan, Chao Lu, Luke E. Berchowitz, Songtao Jia, “The cAMP signaling pathway regulates Epe1 protein levels and heterochromatin assembly” PLOS Genetics (2022).
- Lindsay Moritz et al., “Single residue substitution in protamine 1 disrupts sperm genome packaging and embryonic development in mice” in revision, Nature Structural and Molecular Biology (bioRxiv).
2021
- Saikat Biswas, Joshua D. Karslake, Ziyuan Chen, Ali Farhat, Amanda Ames, Gulzhan Raiymbek, Peter L. Freddolino*, Julie S. Biteen*, and Kaushik Ragunathan*, “HP1 oligomerization compensates for low-affinity H3K9me recognition and provides a tunable mechanism for heterochromatin-specific localization” (bioRxiv).
- Ajay Larkin*, Amanda Ames*, Melissa Seman* and Kaushik Ragunathan, “Investigating mitotic inheritance of histone modifications using tethering strategies” Methods in Molecular Biology, Springer-Nature series: Histone methyltransferases: Methods and Protocols (accepted).
2020
- Raiymbek G, An S, Khurana N, Gopinath S, Larkin, A, Biswas, S, Trievel R, Cho US, Ragunathan K, “An H3K9 methylation dependent protein interaction regulates the non-enzymatic function of a putative histone demethylase” eLife 2020;9:e53155. (PDF) #labinthetimeofcovid-19
2017
- Abeysirigunawardena SC, Kim H, Lai J, Ragunathan, K Rappé MC, Luthey-Schulten Z, Ha T, Woodson SA. “Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes” Nat Commun. 8;8(1):492 (2017).
2015
- Ragunathan, K., G.Jih and D.Moazed “Epigenetic inheritance uncoupled from sequence specific recruitment” Science 348(6230), 1258699 (2015).
2014
- S.Kim , K. Ragunathan , J. Park , C. Joo , D. Kim , and T. Ha “Cooperative conformational transitions keep RecA filament active during ATPase cycle” J. Am. Chem. Soc., 136 (42), pp 14796–14800 (2014).
- Kim, S.C. Abeysirigunawarden, K. Chen, M. Mayerle, K. Ragunathan, Z. Luthey-Schulten, T. Ha , S.A. Woodson, “Protein-guided RNA dynamics during early ribosome assembly”, Nature 506, 334–338 (2014).
News and Views: Kathleen B. Hall “Molecular biology: Protein binding cannot subdue a lively RNA”
2012
- Ragunathan,K., C. Liu and T.Ha, “RecA filament sliding on DNA facilitates homology search”, eLife 1:e00067 (2012).
Commentary – B.Gibb and E.Greene, “Sliding to the rescue of damaged DNA” - H.Koh, M.Kidwell, K. Ragunathan, J.Doudna and S.Myong “ATP-independent diffusion of double stranded RNA binding proteins”, Proc.Natl.Acad.Sci 110(1) 151-156 (2012).
2011
- Ragunathan,K., C. Joo and T.Ha, “Real time observation of strand exchange reaction with high spatiotemporal resolution”, Structure 19(8) 1064-73 (2011).
Commentary – C.Wyman, “Mechanistic insight from Chaos: How RecA Mediates DNA Strand Exchange”. Evaluated at F1000 biology - Jain, A., R.Liu, B.Ramani, E.Aruaz, Y.Ishitsuka, K. Ragunathan, J. Park, J. Chen, Y. Xiang and T.Ha, “Probing cellular protein complexes using single-molecule pull-down”, Nature 473, 484-488 (2011).
2010
- Lee, J., S. Lee, K. Ragunathan, C. Joo, T. Ha and S. Hohng, “Single-molecule four-color FRET”, Angew. Chem. Int. Ed. 49(51), 9922-9925 (2010).