Piali Sengupta
 Harold and Bernice Davis Chair in Aging and Neurodegenerative Disease, and Professor of Biology
Harold and Bernice Davis Chair in Aging and Neurodegenerative Disease, and Professor of Biology
Research Description
Neurogenetics of sensory behaviors
How do animals sense their environment? How do these external cues elicit specific behaviors and developmental programs? How are these responses modified by the animal’s experience and internal state? The lab aims to answer these questions by identifying the genetic, molecular, and neuronal mechanisms that allow us to robustly, yet flexibly, sense and respond to our complex and constantly changing environment. Understanding the regulation of sensory signaling and signal processing is of significant biomedical importance since disruption of these pathways leads to a range of neurological and behavioral disorders.
To address the above issues, we use a multifaceted approach combining genetic, genomic, and molecular tools, high-resolution quantitative behavioral assays, live imaging, and in vivo analyses of neuronal activity. Our experimental organism is C. elegans, but we also work with mammalian primary neurons and cultured cells. We greatly value collaborations and open communication, and seek out collaborative opportunities that allow us to learn from others, and build new scientific connections within and across disciplines.
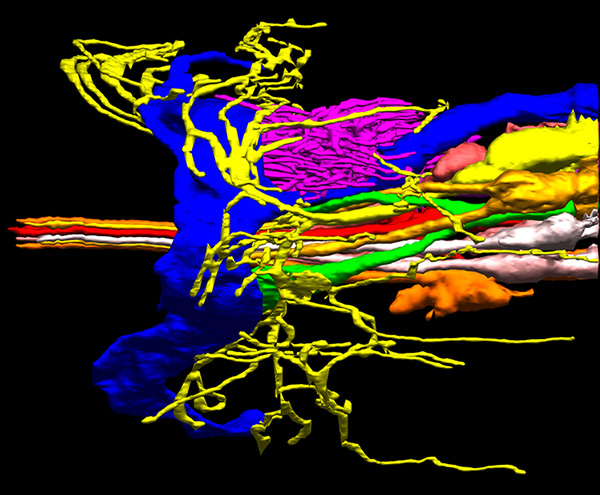 Sensory neuron cilia in C. elegans. From Doroquez et al., eLife 2014.
Sensory neuron cilia in C. elegans. From Doroquez et al., eLife 2014.
Ongoing work in the lab can be broadly grouped into two areas.
- C. elegans exhibits remarkably sensitive responses to attractive and aversive chemicals in its environment, as well as to temperature changes. For instance, worms can detect temperature changes of as little as 0.01°C over a >10°C temperature range. These responses are modulated by the animal’s past experience, current context, and satiety state. The ‘Axis of Taxis’ in the lab identifies molecules required for chemo- and thermosensory signal transduction, describes neuronal responses to chemical and thermal cues and how these responses are modulated by experience and internal state, and investigates the sensorimotor transformations that translate these stimuli into specific behavioral programs.
- All sensory signaling molecules in sensory neurons are localized to their cilia, microtubule-based structures that are specialized to concentrate components of signaling cascades, and that represent the site of primary signal transduction. The morphologies of sensory cilia are remarkably complex and diverse. The ‘Cilia Squad’ in the lab investigates how different types of cilia are generated and maintained, how signaling proteins are trafficked and localized to cilia, how unique cilia morphologies dictate sensory responses, and how sensory activity in turn reshapes cilia architecture.
Please see the lab website and our publications for additional details.
Selected Publications
- Context-dependent inversion of the response in a single sensory neuron type reverses olfactory preference behavior. Khan M, Hartmann AH, O'Donnell MP, Piccione M, Chao PH, Dwyer N.D., Bargmann C. I., Sengupta P. bioRxiv, 2021 Nov 9. doi: 10.1101/2021.11.08.467792
- xbx-4, a homolog of the Joubert syndrome gene FAM149B1, acts via the CCRK and RCK kinase cascade to regulate cilia morphology. Maurya AK, Sengupta P. Current Biology. 2021 Nov 2. doi: 10.1016/j.cub.2021.10.027
- Chemosensory signal transduction in Caenorhabditis elegans. Ferkey D, Sengupta P, L'Etoile Noelle D. Genetics. 2021 Mar 31. doi: 10.1093/genetics/iyab004
- Ciliary neuropeptidergic signaling dynamically regulates excitatory synapses in postnatal neocortical pyramidal neurons. Tereshko L, Gao Y, Cary BA, Turrigiano G, Sengupta P. eLife. 2021 Mar 2. doi: 10.7554/eLife.65427
- Chronic vs acute manipulations reveal degeneracy in a thermosensory neuron network. Yeon J, Takeishi A, Sengupta P. MicroPubl Biol. 2021 Jan 15. doi: 10.17912/micropub.biology.000355
- GRDN-1/Girdin regulates dendrite morphogenesis and cilium position in two specialized sensory neuron types in C. elegans. Nechipurenko I, Lavrentyeva S, Sengupta P. Dev Biol. 2021 Jan 16. doi: 10.1016/j.ydbio.2020.12.022
- Feeding state functionally reconfigures a sensory circuit to drive thermosensory behavioral plasticity. Takeishi A, Yeon J, Harris N, Yang W, Sengupta P. eLife. 2020 Oct 19. doi: 10.7554/eLife.61167
- The URX oxygen-sensing neurons in C.elegans are ciliated. Kazatskaya A, Yuan L, Amin-Wetzel N, Philbrook A, de Bono M, Sengupta P. microPublication Biology. 2020 Sep 20. doi: 10.17912/micropub.biology.000303.
- A neurotransmitter produced by gut bacteria modulates host sensory behaviour. O'Donnell MP, Fox BW, Chao PH, Schroeder FC, Sengupta P. Nature. 2020 Jun 17. doi: 10.1038/s41586-020-2395-5.
- The C. elegans Tubby homolog dynamically modulates olfactory cilia membrane morphogenesis and phospholipid composition. DiTirro, D, Philbrook, A, Rubino, K, & Sengupta, P. eLife. 2019 Jul 1. doi: 10.7554/eLife.48789
- How Caenorhabditis elegans Senses Mechanical Stress, Temperature, and Other Physical Stimuli. Goodman MB, Sengupta P. Genetics. 2019 May;212(1):25-51. doi: 10.1534/genetics.118.300241.
- A CCRK and a MAK Kinase Modulate Cilia Branching and Length via Regulation of Axonemal Microtubule Dynamics in Caenorhabditis elegans. Maurya AK, Rogers T, Sengupta P. Curr Biol. 2019 Apr 22;29(8):1286-1300.e4. doi: 10.1016/j.cub.2019.02.062.
- Thermosensation: Human Parasitic Nematodes Use Heat to Hunt Hosts. O'Donnell MP, Khan M, Sengupta P. Curr Biol. 2018 Jul 23;28(14):R795-R798. doi: 10.1016/j.cub.2018.05.082.
- The extraordinary AFD thermosensor of C. elegans. Goodman MB, Sengupta P. Pflugers Arch. 2018 May;470(5):839-849. doi: 10.1007/s00424-017-2089-5.
- Rictor/TORC2 mediates gut-to-brain signaling in the regulation of phenotypic plasticity in C. elegans. O'Donnell MP, Chao PH, Kammenga JE, Sengupta P. PLoS Genet. 2018 Feb 7;14(2):e1007213. doi: 10.1371/journal.pgen.1007213. eCollection 2018 Feb.
- Centriolar remodeling underlies basal body maturation during ciliogenesis in Caenorhabditis elegans. Nechipurenko IV, Berciu C, Sengupta P, Nicastro D. Elife. 2017 Apr 15;6. pii: e25686. doi: 10.7554/eLife.25686.
- Structural and Functional Recovery of Sensory Cilia in C. elegans IFT Mutants upon Aging. Cornils A, Maurya AK, Tereshko L, Kennedy J, Brear AG, Prahlad V, Blacque OE, Sengupta P. PLoS Genet. 2016 Dec 1;12(12):e1006325. doi: 10.1371/journal.pgen.1006325. eCollection 2016 Dec.
- A Conserved Role for Girdin in Basal Body Positioning and Ciliogenesis. Nechipurenko IV, Olivier-Mason A, Kazatskaya A, Kennedy J, McLachlan IG, Heiman MG, Blacque OE, Sengupta P. Dev Cell. 2016 Sep 12;38(5):493-506. doi: 10.1016/j.devcel.2016.07.013.
- Receptor-type Guanylyl Cyclases Confer Thermosensory Responses in C. elegans. Takeishi A, Yu YV, Hapiak VM, Bell HW, O'Leary T, Sengupta P. Neuron. 2016 Apr 20;90(2):235-44. doi: 10.1016/j.neuron.2016.03.002.
- Feeding state-dependent regulation of developmental plasticity via CaMKI and neuroendocrine signaling. Neal SJ, Takeishi A, O'Donnell MP, Park J, Hong M, Butcher RA, Kim K, Sengupta P. Elife. 2015 Sep 3;4. pii: e10110. doi: 10.7554/eLife.10110. Erratum in: Elife. 2015;4:e11547. PMID: 26335407
- CaMKI-dependent regulation of sensory gene expression mediates experience-dependent plasticity in the operating range of a thermosensory neuron. Yu YV, Bell HW, Glauser D, Van Hooser SD, Goodman MB, Sengupta P. Neuron. 2014 Dec 3;84(5):919-926. doi: 0.1016/j.neuron.2014.10.046. PMID: 25467978
- A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Doroquez DB*, Berciu C*, Anderson JR, Sengupta P*, Nicastro D.* Elife. 2014 Mar 25;3:e01948. doi: 10.7554/eLife.01948. PMID: 24668170
- Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Jang H*, Kim K*, Neal SJ, Macosko E, Kim D, Butcher RA, Zeiger DM, Bargmann CI*, Sengupta P.* Neuron. 2012 Aug 23;75(4):585-92. doi: 10.1016/j.neuron.2012.06.034.PMID: 22920251
* - co-corresponding authors