Capsid Assembly Around a Polymer
We performed the first ever simulations of simultaneous capsid assembly and encapsulation of a polymer, we characterized the role of the polymer in this cooperative assembly process and obtained experimentally testable predictions about assembly failures that occur in the presence of long polymers.
In particular, we identified relationships between experimental control parameters (e.g. polymer length, protein concentration, and solution conditions) and assembly morphologies. As the polymer length increases beyond its optimal length, the simulations predict the formation of multiplet capsids which share a single polymer (Fig. 1, lower right). Concurrent experiments by the group of C. Knobler and W. Gelbart (UCLA), in which capsid proteins assemble around RNA molecules with lengths in excess of the viral genome length, resulted in similar structures. [1, 2]
Our computational and theoretical modeling demonstrated that there are two classes of assembly pathways or mechanisms which can occur around a central core (i.e., RNA or a polymer), each of which leads to a different assembly kinetics. Fig. 2 shows snapshots from two trajectories, each of which provides an example of one of the two classes. In the trajectory shown in Fig. 2A, the capsid assembles by a nucleation and growth mechanism. First a small partial capsid nucleates on the polymer, after which one or a few subunits at a time reversibly add until the capsid is completed. In the second mechanism (Fig. 2B) the subunits first rapidly adsorb onto the polymer in disordered configurations followed by cooperative rearrangement and unbinding of some subunits to result in an ordered capsid structure. Our simulations and theory predict that the assembly mechanism can be tuned experimentally by changing the surface charge density or solution conditions [1, 3, 4]. The simulations also show that the polymer has a substantial effect on the thermodynamics and kinetics of the assembly process. Most importantly, interactions with the polymer stabilize the partially growing capsid, substantially reducing the nucleation barrier [2]. Secondly, the polymer dramatically increases the flux of incoming subunits through two mechanisms. First, adsorbed subunits are brought to the edge of the growing capsid by polymer looping motions, much like the action of a fly caster or the Cookie Monster. Secondly, subunits can adsorb onto the polymer and slide via one-dimensional diffusion or hopping until reaching the edge of the growing capsid. This mechanism was first predicted by Hu and Schlovskii [5] and then seen in action in our molecular simulations [1, 2].
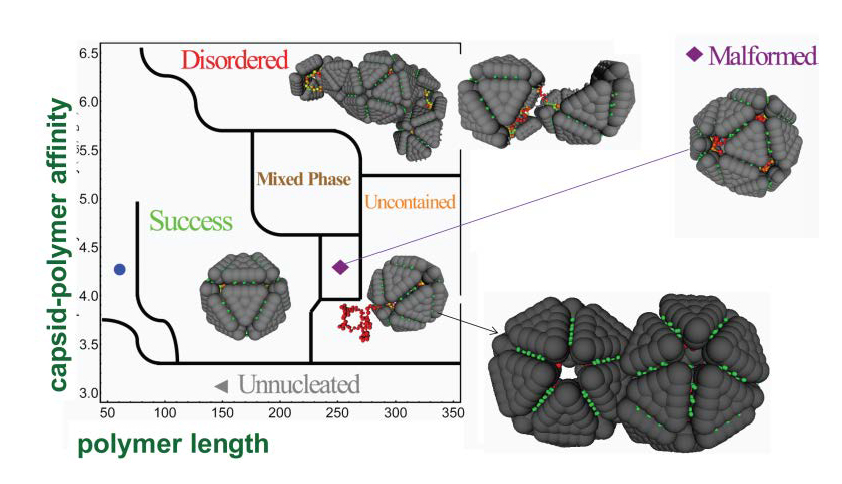
Fig. 1: Results from simulations of model capsid proteins assembling around a flexible polymer. The simulation predictions for the dominant assembly product are shown as a function of polymer length and the strength of polymer-protein interactions (controlled by salt concentration). The structure at the lower right which forms for long polymers was recently observed in experiments. Figure is from [1].
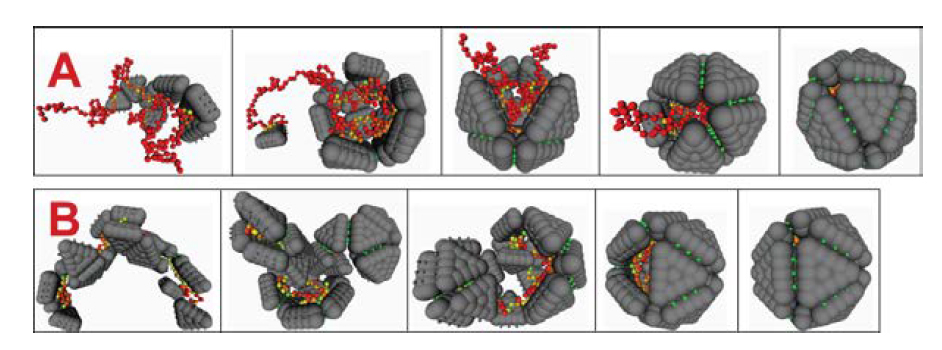
Fig. 2: Two mechanisms for assembly around a polymer. (A) A small partial capsid nucleates on the polymer, followed by the reversible addition of one or twosubunits at a time. Cooperative polymer-subunit motions enhance the flux of incoming subunits. (B) Subunits adsorb onto the polymer in a disordered manner, followed by cooperative annealing and unbinding of excess subunits. Figure from [1].
References
- Elrad, O.M. and M.F. Hagan, "Encapsulation of a Polymer by an Icosahedral Virus." Physical Biology, 2010. 7: p. 045003.
- Kivenson, A. and M.F. Hagan, "Mechanisms of Capsid Assembly Around a Polymer." Biophysical Journal, 2010. 99: p. 619.
- Hagan, M.F., "A Theory for Viral Capsid Assembly Around Electrostatic Cores." Journal of Chemical Physics, 2009. 130: p. 114902.
- Hagan, M.F., "Controlling Viral Capsid Assembly With Templating." Physical Review E, 2008. 77(5).
- Hu, T. and B.I. Shklovskii, "Kinetics of Viral Self-Assembly: Role of the Single-Stranded RNA Antenna." Physical Review E, 2007. 75: p. 051901.