Size Control and Polymorphism Mechanisms in Capsid Assembly
Many pathogenic viruses require a specific structure to be infectious, and their proteins form that structure in vivo with remarkable fidelity.
The same proteins, however, assemble into capsids with different sizes and morphologies, still with high selectivity, to accommodate nucleic acids [1], inorganic nanoparticles [2] and polyanions [3, 4] with different sizes. For example, Fig. 1 shows electron microscopy images of brome mosaic virus (BMV) proteins assembled into icosahedral morphologies with different numbers of capsid proteins around charge-functionalized nanoparticles with commensurate sizes. What features of viral components enable assembly that is so selective and yet also highly adaptable?
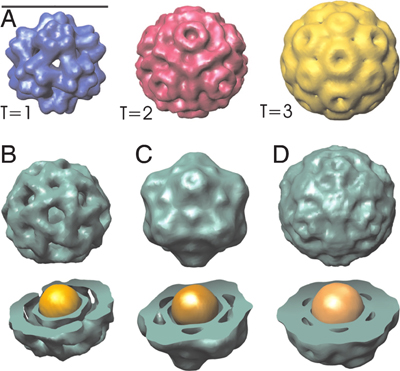 Figure 1: Electron microscopy images of brome mosaic virus (BMV) capsid proteins assembled around charge-functionalized nanoparticles with diameters: (B) 6 nm, (C) 9 nm, and (D) 12 nm. In the top panel (A), images of empty capsids are shown with T=1 (60 proteins), pseudo T=2 (120 proteins), and T=3 (180 proteins) morphologies. Figure taken from Ref. [2].
Figure 1: Electron microscopy images of brome mosaic virus (BMV) capsid proteins assembled around charge-functionalized nanoparticles with diameters: (B) 6 nm, (C) 9 nm, and (D) 12 nm. In the top panel (A), images of empty capsids are shown with T=1 (60 proteins), pseudo T=2 (120 proteins), and T=3 (180 proteins) morphologies. Figure taken from Ref. [2].
Background: The Geometry of Spherical Capsids
As first suggested by Watson and Crick [5], the subunits of many spherical viruses are arranged with icosahedral symmetry. At most 60 identical subunits, however, can be arranged with perfect icosahedral symmetry, while structural experiments indicated that many capsids have multiples of 60 subunits. Caspar and Klug proposed geometrical arguments that describe how multiples of 60 proteins can be arranged with icosahedral symmetry, where individual proteins interact through the same interfaces but take slightly different, or quasi-equivalent, conformations [6]. Protein subunits can be grouped into morphological units, usually as pentamers and hexamers. Icosahedral symmetry requires exactly 12 pentamers, located at the vertices of an icosahedron inscribed within the capsid. A complete capsid is comprised of 60T subunits, where T is the "triangulation number", which is equal to the number of distinct subunit conformations. For example, images of the x-ray structures for T=3 and T=1 BMV capsids are shown in Fig. 2; the three different subunit conformations are differentiated by color. The principles of quasi-equivalence are reviewed with exceptional clarity in Ref. [7].
The dynamical process by which protein conformations are "selected" during the assembly process is not well understood because the structures of transient assembly intermediates have not been accessible to experiments. Berger and coworkers [8] showed that well-formed capsids can result if assembly follows "local rules", in which only subunits with the conformation dictated by adjacent subunits can bind to and assembling capsid. For many viruses, however, subunit-subunit binding interfaces show little variation between different conformations in the crystal structure, and thus it is not clear how conformation-specific subunit-subunit binding interactions can be. Furthermore, how can BMV proteins that assemble with conformational specificity to form T=3 empty capsids adapt to form T=1 and pseudo-T=2 capsids around commensurate cargoes?
Modeling Capsid Assembly and Polymorphism
In Ref. [9], we describe simulations of a simplified model (Fig. 3), derived from the crystal structures of BMV T=3 and T=1 capsids, that describes the assembly of icosahedral capsids from subunits that interconvert between different conformations (or quasi-equivalent states). We systematically vary the extent to which binding between subunits depends on conformation and the relative probabilities for subunits to take different conformations. We find that even weakly conformation-specific subunit-subunit binding interactions enable robust assembly of T=3 empty capsids. Specifically, the binding free energy for the conformation pairs seen in the crystal structure only needs to be more favorable than other conformation pairs by as much as the thermal energy (kBT). Secondly, we find a narrow, but physically reasonable, range of system parameters for which only T=3 empty capsids assemble, but T=1 capsids form on a commensurate nanoparticle. Assembly that is adaptive to the size of the cargo, however, requires moderate cargo-subunit interaction strengths. Strong nanoparticle-subunit interactions frustrate nanoparticle encapsidation by stabilizing partial capsids with curvature that is incommensurate with the nanoparticle size (Fig. 4). For details please see Refs. [9], [10], and [11].
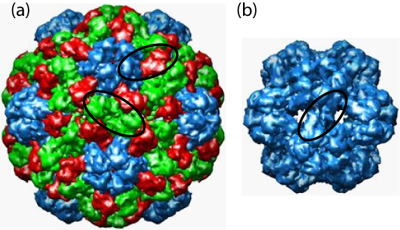 Figure 2: Images of brome mosaic virus (BMV) capsids with (a) 180 subunits (T=3) and (b) 60 subunits (T=1). Subunits with quasi-equivalent conformations (defined in the text) A, B, and C are blue, red, and green, respectively. The black ovals outline examples of the protein dimers, related by a twofold or quasi-twofold axis of symmetry, that are believed to be the basic assembly unit. Note that the only dimers present in the crystal structures are AB and CC for T=3 and AA for T=1. Capsid images are from the VIPER webpage [12].
Figure 2: Images of brome mosaic virus (BMV) capsids with (a) 180 subunits (T=3) and (b) 60 subunits (T=1). Subunits with quasi-equivalent conformations (defined in the text) A, B, and C are blue, red, and green, respectively. The black ovals outline examples of the protein dimers, related by a twofold or quasi-twofold axis of symmetry, that are believed to be the basic assembly unit. Note that the only dimers present in the crystal structures are AB and CC for T=3 and AA for T=1. Capsid images are from the VIPER webpage [12].
A.
B.
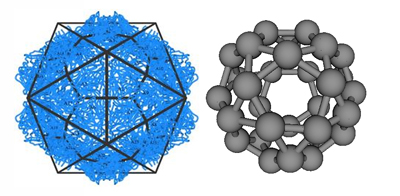
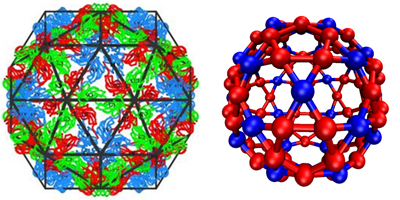 Figure 3: Model capsid geometries. (left) Images of BMV capsid crystal structures with (a) T=1 and (b) T=3 morphologies, overlaid with icosahedral cages. Subunits with quasi-equivalent conformations A, B, and C are blue, red, and green, respectively. (Right) model capsid geometries. Each subunit represents a protein dimer (the basic assembly unit for BMV). Subunit sizes are reduced to aid visibility, and colors indicate that conformations of proteins within a dimer subunit: Silver, AA; red, AB; blue, CC.
Figure 3: Model capsid geometries. (left) Images of BMV capsid crystal structures with (a) T=1 and (b) T=3 morphologies, overlaid with icosahedral cages. Subunits with quasi-equivalent conformations A, B, and C are blue, red, and green, respectively. (Right) model capsid geometries. Each subunit represents a protein dimer (the basic assembly unit for BMV). Subunit sizes are reduced to aid visibility, and colors indicate that conformations of proteins within a dimer subunit: Silver, AA; red, AB; blue, CC.
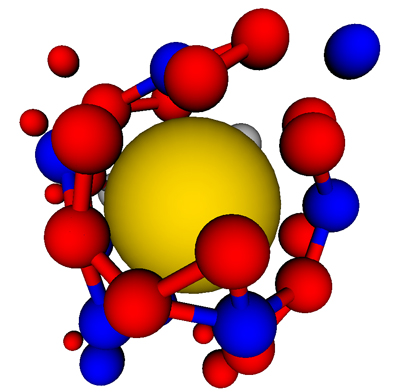
Figure 4: (A) A frustrated partial capsid with a T=3 morphology grows on a T=1-sized nanoparticle. The subunits near the nanoparticle are stabilized by its attraction, but the mismatch between the capsid and nanoparticle geometry disfavors the addition of nanoparticles.
;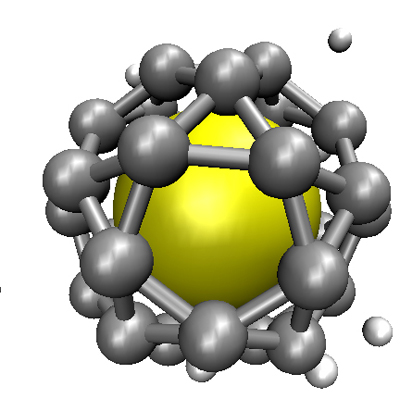
(B) If subunit-nanoparticle are not too strong, the metastable partial capsid can disassemble, enabling a T=1 capsid to nucleate and grow to completion.
References
-
Krol, M. A., et al., RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proceedings of the National Academy of Sciences of the United States of America, 1999. 96(24): p. 13650-13655.
-
Sun, J., et al., Core-controlled polymorphism in virus-like particles. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104(4): p. 1354-1359.
-
Bancroft, J. B., E. Hiebert, and C. E. Bracker, Effects of Various Polyanions on Shell Formation of Some Spherical Viruses. Virology, 1969. 39(4): p. 924-&.
-
Hu, Y., et al. Packaging of a polymer by viral nanocontainer. in American Institute of Chemical Engineers Annual Meeting. 2007. Salt Lake City, Utah.
-
Crick, F. H. C. and J. D. Watson, Structure of Small Viruses. Nature, 1956. 177(4506): p. 473-475.
-
Caspar, D. L. D. and A. Klug, Physical Principles in Construction of Regular Viruses. Cold Spring Harbor Symposia on Quantitative Biology, 1962. 27: p. 1-&.
-
Johnson, J. E. and J. A. Speir, Quasi-equivalent viruses: A paradigm for protein assemblies. Journal of Molecular Biology, 1997. 269(5): p. 665-675.
-
Berger, B., et al., Local Rule-Based Theory of Virus Shell Assembly. Proceedings of the National Academy of Sciences of the United States of America, 1994. 91(16): p. 7732-7736.
-
Elrad, O. M. and M.F. Hagan, Mechanisms of Size Control and Polymorphism in Viral Capsid Assembly. Nano Letters, 2008. 8(11): p. 3850-3857.
-
Hagan, M. F., Controlling Viral Capsid Assembly with Templating. Phys. Rev. E (in press), arxiv:0712.3786v1 [q-bio.BM], 2008.
-
Hagan, M. F., A theory for viral capsid assembly around electrostatic cores. arXiv:0808.2204v1 [q-bio.BM], 2008.
-
Reddy, V. S., et al., Virus Particle Explorer (VIPER), a Website for virus capsid structures and their computational analyses. Journal of Virology, 2001. 75(24): p. 11943-11947.