Niels Bradshaw
 Assistant Professor of Biochemistry
Assistant Professor of Biochemistry
Research Description
Regulation of Protein Phosphatases and the Evolution of Cellular Signaling
Phosphatases and kinases integrate diverse cellular and environmental cues to precisely and rapidly tune the activity of target proteins by controlling their phosphorylation state. We use genetics, cell biology, enzymology and structural biology to determine how protein phosphatases achieve regulation, specificity, and evolutionary flexibility.
The PP2C family of protein serine/threonine phosphatases is widespread (found throughout the bacterial, plant, and animal kingdoms), highly diversified (some species of bacteria and plants have more than 50 and humans have 20), and has broad impact on biology (controlling diverse processes including stress responses, development, and cell growth and death).
How are phosphatases regulated?
To accomplish their diverse functions, PP2C phosphatases have divergent cis-regulatory domains that receive cellular signals and control phosphatase activity. I discovered a conserved switch within the PP2C phosphatase domain that controls activity by recruiting a divalent metal cofactor in response to regulatory domain movement. A major future goal is to understand how this switch is exploited by diverse phosphatases to respond to different signals and execute crucial biological decisions.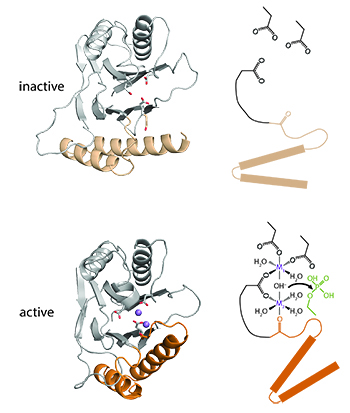 What do phosphatases sense?
What do phosphatases sense?
The most widespread role of PP2C phosphatases is control of the general stress response in bacteria, which is important for virulence, antibiotic resistance, and persistence. However, the signals phosphatases directly sense to initiate the general stress response are virtually unknown. PP2C phosphatases in humans and other metazoans similarly regulate critical cellular decisions in response to largely unidentified signals. We pursue identification of such signals by a combined genetic and biochemical strategy. The signals an organism uses to detect stress indicate what is important for survival of that organism in the environment and within communities. These signals will also suggest targets for species-specific antimicrobials and other targeted therapies.
How do phosphatases achieve specificity?
PP2C phosphatases are highly substrate specific, which is critical to their biological functions. In contrast to kinases, which have a substrate-binding pocket, the phosphatase active site is exposed on the surface of the protein. How this exposed active site is restricted to acting on cognate substrates is unknown. I hypothesize that the regulatory switch facilitates substrate discrimination by integrating binding of cognate substrate with activation by regulatory inputs.
How have phosphatases evolved and diversified?
The modularity of PP2C phosphatase regulation has conferred evolutionary adaptability, allowing a common enzyme core to be connected to new regulatory inputs and substrates. How has this regulatory mechanism been adapted by evolution to diverse functions? The identification of the PP2C regulatory switch additionally revealed that PP2C phosphatases and a family of proteases (including the proteasomal proteases) are structurally related, and both use this regulatory switch to control their unrelated catalytic mechanisms. This suggests that allosteric regulatory mechanisms can facilitate the evolution of new catalytic activities and establishes PP2C phosphatases and proteasomal proteases as an experimental system with which to study how such evolution occurs. What evolutionary pathway might accomplish such a change? One possibility is that an intermediate existed that did not perform either chemistry but preserved the regulatory mechanism. Indeed, pseudo-PP2C phosphatases that have lost their catalytic activities perform non-catalytic regulatory activities in diverse organisms.
Selected Publications
-
Bradshaw, N., Levdikov, V.M., Zimanyi, C.M., Gaudet, R., Wilkinson, A.J., and Losick, R. (2017) A widespread family of serine/threonine protein phosphatases shares a common regulatory switch with proteasomal proteases. eLife, 6:e26111
-
Bradshaw, N. & Losick, R. (2015). Asymmetric division triggers cell-specific gene expression through coupled capture and stabilization of a phosphatase. eLife, 4:e08145.
-
DeFrancesco, A.S., Masloboeva, N., Syed, A.K., Deloughery, A., Bradshaw, N., Li, G.-W., Gilmore, M.S., Walker, S., and Losick, R. (2017). Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci USA10.1073/pnas.1704544114.
-
Subramaniam, A. R., Deloughery, A., Bradshaw, N., Chen, Y., O'Shea, E., Losick, R., & Chai, Y. (2013). A serine sensor for multicellularity in a bacterium. eLife 2:e01501.
-
Bradshaw N.*, Neher S. B.*, Booth D. S., Walter P. (2009) Signal sequences activate the catalytic switch of SRP RNA. Science323(5910):127-130.
-
Neher S. B.*, Bradshaw N.*, Floor S. N., Gross J. D., Walter P. (2008) SRP RNA controls a conformational switch regulating the SRP-SR interaction. Nat Struct Mol Biol 15:916-923.
-
Bradshaw N., Walter P. (2007). Signal recognition particle (SRP) RNA links conformational changes in the SRP to protein targeting. Mol Biol Cell 7:2728-34.