Jeff Gelles

Aron and Imre Tauber Professor of Biochemistry and Molecular Pharmacology
Research Description
Single-molecule Biochemistry and Biophysics. Transcription and RNA processing. Cytoskeletal networks and regulation.
Living cells are chock full of dynamic complexes of protein, RNA, and DNA molecules. We want to understand the fundamental chemical and physical mechanisms through which these molecular machines perform essential biological processes. Our research is focused in two different areas of study: 1) the function of the molecular machines essential to gene expression and its regulation, in particular those that control the synthesis and processing of messenger RNAs, and 2) the function of molecular machines essential to the organization and utilization of the actin and microtubule cytoskeleton of eukaryotic cells.
All of the processes that we study involve dynamic molecular assemblies and multiple reaction intermediates. It is challenging to study their mechanisms using conventional biochemical approaches because different individual molecules in a population are doing different things at the same time. To overcome this “crowd noise problem”, we have developed and used single-molecule light microscopy methods that allow us to observe the behavior of isolated individual molecules and molecular complexes in real time. Using these techniques we can directly observe the assembly, rearrangements, and disassembly of molecular complexes, characterize conformational changes, and detect biochemical reactions. Together, these methods permit comprehensive investigation of molecular mechanisms, revealing reaction pathways and allowing quantitative evaluation and modeling of reaction dynamics. We perform these single-molecule experiments both in systems reconstituted from purified componentsand in extracts that recapitulate the molecular complexity of the living cell.

Our research uses approaches from scientific fields ranging from cell biology and genetics through biochemistry and molecular biology to physical chemistry and condensed-matter physics. In our experience, the most exciting advances in science often arise when scientists from different disciplines collaborate.
Selected Publications
- Stumper SK, Ravi H, Friedman LJ, Mooney RA, Corrêa IR, Gershenson A, Landick R, Gelles J. Delayed inhibition mechanism for secondary channel factor regulation of ribosomal RNA transcription. eLife. 2019; 8.
- Guo S, Sokolova OS, Chung J, Padrick S, Gelles J, Goode BL. Abp1 promotes Arp2/3 complex-dependent actin nucleation and stabilizes branch junctions by antagonizing GMF. Nature communications. 2018; 9(1):2895.
- Braun JE, Friedman LJ, Gelles J, Moore MJ. Synergistic assembly of human pre-spliceosomes across introns and exons. eLife. 2018; 7.
- Chadda R, Krishnamani V, Mersch K, Wong J, Brimberry M, Chadda A, Kolmakova-Partensky L, Friedman LJ, Gelles J, Robertson JL. The dimerization equilibrium of a ClC Cl(-)/H(+) antiporter in lipid bilayers. eLife. 2016; 5.
- Friedman LJ, Gelles J. Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods (San Diego, Calif.). 2015; 86:27-36.
- Ticau S, Friedman LJ, Champasa K, Corrêa IR Jr, Gelles J, Bell SP. (2017) "Mechanism and timing of Mcm2-7 ring closure during DNA replication origin licensing." Nature structural & molecular biology. 2017; 24(3):309-315.
- Tetone LE, Friedman LJ, Osborne ML, Ravi H, Kyzer S, Stumper SK, Mooney RA, Landick R, Gelles J. (2017) "Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue." Proceedings of the National Academy of Sciences of the United States of America. 2017; 114(7):E1081-E1090.
- Hoskins AA, Rodgers ML, Friedman LJ, Gelles J, Moore MJ. (2016) "Single molecule analysis reveals reversible and irreversible steps during spliceosome activation." eLife (2016) 5:e14166.
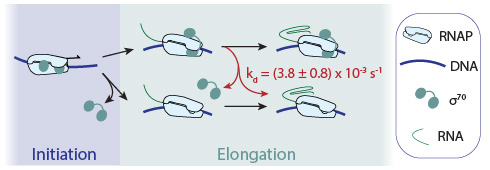
- Harden TT, Wells CD, Friedman LJ, Landick R, Hochschild A, Kondev J, Gelles J. (2016) "Bacterial RNA polymerase can retain σ70 throughout transcription." Proceedings of the National Academy of Sciences of the United States of America. 2016; 113(3):602-7.</a
- Bombardier JP, Eskin JA, Jaiswal R, Corrêa IR Jr, Xu MQ, Goode BL, Gelles J. (2015) "Single-molecule visualization of a formin-capping protein 'decision complex' at the actin filament barbed end." Nature Communications. 2015 Nov 13;6:8707.
- Jansen S, Collins A, Chin SM, Ydenberg CA, Gelles J, Goode BL. (2015) "Single-molecule imaging of a three-component ordered actin disassembly mechanism." Nature Communications 2015 May 21;6:7202.
- Paramanathan T, Reeves D, Friedman LJ, Kondev J, Gelles J. (2014) "A general mechanism for competitor-induced dissociation of molecular complexes." Nature Communications. 2014; 5:5207.
- Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Corrêa IR Jr, Xu MQ, Gelles J, Moore MJ. (2013) "Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy." Cell Reports. 2013 Oct 17;5(1):151-65.
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Corrêa IR Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. (2013) "Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation." Elife. 2013 Sep 3;2:e01008.
- Friedman LJ, Mumm JP, Gelles J.(2013) "RNA polymerase approaches its promoter without long-range sliding along DNA." Proceedings of the National Academy of Sciences of the United States of America. 2013; 110(24):9740-5.
- Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. (2013) "Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5' splice site and branch site." Proceedings of the National Academy of Sciences of the United States of America. 2013; 110(17):6783-8.
- Smith BA, Daugherty-Clarke K, Goode BL, Gelles J.(2013) "Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging." Proceedings of the National Academy of Sciences of the United States of America.2013 Jan 22;110(4):1285-90. 2013; 110(4):1285-90.