Julia Kardon
 Assistant Professor of Biochemistry
Assistant Professor of Biochemistry
Research Description
Mechanisms for control of mitochondrial protein activity, quality, and lifespan
The activity and quality of mitochondrial proteins must be controlled to match the dynamic needs of eukaryotic organisms. This control is accomplished both by a chaperone machinery (related to the ancestral bacterial endosymbiont chaperones) and by degradation of mitochondrial subcompartments by the lysosome. How proteins are specifically directed to these machineries and how the outcome of their interaction is determined remains mysterious. I employ diverse biochemical and biophysical approaches to delineate the molecular mechanisms of this control, in combination with cell biological, genetic, and proteomic tools to discover new features and components of these systems.
How is unfolding targeted for mitochondrial protein biogenesis, repair, and degradation?
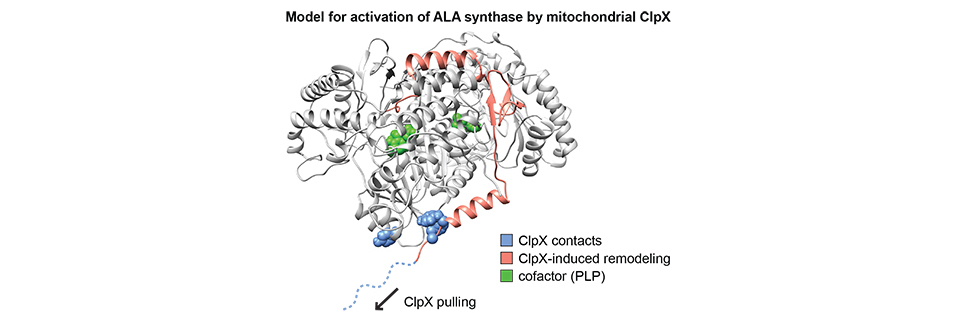
How mitochondrial chaperones interpret signals in their substrate proteins and the cellular environment to control the activities of these proteins is a major outstanding question. I discovered a conserved function for the mitochondrial protein unfoldase ClpX as an activator of the initial enzyme in heme biosynthesis, ALA synthase. ClpX performs an unusually targeted, partial unfolding of ALA synthase that facilitates cofactor binding. My investigation of a newly discovered erythropoietic disease allele of ClpX indicates that ClpX may conditionally direct degradation of ALAS as well. The ClpX-ALAS chaperone-substrate pair thus provides a system for biochemical and structural investigation of several mechanisms for control of mitochondrial protein activity as well as general principles that determine the outcome of chaperone-substrate interactions, from remodeling to complete unfolding. These variable outcomes allow ClpX and other chaperones to accomplish a diverse array of functions in the cell.
How does ClpX adapt the mitochondrial proteome to cellular demand and stress?
Loss or dysregulation of mitochondrial ClpX or ClpP results in mitochondrial dysfunction and human diseases that suggest multiple important functions beyond heme biosynthesis. These loss-of-function phenotypes do not include general defects in protein quality control, indicating that like its better-characterized bacterial homolog, mitochondrial ClpX is specialized for regulatory and conditional action on a specific repertoire of client proteins. My elucidation of ClpX activation of ALAS also highlighted its dual ability to remodel or degrade its substrates. I am investigating the signals that direct substrate and condition-specific protein unfolding by ClpX. Understanding the mechanism of substrate selection will reveal how the mitochondrion makes specific and crucial regulatory decisions.
How are mitochondrial proteins selected for lysosomal degradation?
In addition to regulation and degradation within the mitochondrion, mitochondrial proteins can be degraded by transport of mitochondrial subcompartments to the lysosome (through mitophagy or vesicular transport). Selectivity for protein identity and/or quality in degradation-marked subcompartments has been observed in diverse systems. The physical basis of this selectivity is undetermined, as is the functional differentiation between mitochondrial-autonomous and lysosomal strategies for protein degradation.
Selected Publications
-
Kardon, J. R., Moroco, J. A., Engen, J. R., Baker, T. A. (2018) Mitochondrial ClpX activates an essential metabolic enzyme through partial unfolding. bioRxivdoi: 10.1101/327056
-
Brown, B. L., Kardon, J. R., Sauer, R. T., Baker, T. A. (2018) Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme. Structure 26(4):580-589
-
Yien, Y. Y., Ducamp, S., van der Vorm, L. N., *Kardon, J. R., Manceau, H., Kannengiesser, C., Bergonia, H. A., Kafina, M. D., Karim, Z., Gouya, L., Baker, T. A., Puy, H., Phillips, J. D., Nicolas, G. & Paw, B. H. (2017) Mutation in human CLPX elevates levels of δ-aminolevulinate synthase and protoporphyrin IX to promote erythropoietic protoporphyria. PNAS 114:E8045–E8052 *co-second author
-
Kardon, J. R., Yien, Y. Y., Huston, N. C., Branco, D. S., Hildick-Smith, G. J., Rhee, K. Y., Paw, B. H., Baker, T. A. (2015) Mitochondrial ClpX activates a key enzyme for heme biosynthesis and erythropoiesis. Cell 161(4):858-67
-
Al-Furoukh, N., Kardon, J. R., Krüger, M., Szibor, M., Baker, T. A., Braun, T. (2014) NOA1, a novel ClpXP substrate, takes an unexpected nuclear detour prior to mitochondrial import. PLoS ONE 9(7):e103141
-
Kardon, J. R., Vale, R. D. (2009) Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 10(12):854-65
-
Kardon, J. R., Reck-Peterson, S. L., and Vale, R. D. (2009) Regulation of dynein processivity and cellular function by dynactin. PNAS106:5669-74