Douglas Theobald
 Professor of Biochemistry
Professor of Biochemistry
Department Chair, Biochemistry
Fields of Specialization
-
Structure and function of single-stranded nucleic acid protein complexes
-
Likelihood and Bayesian techniques in structural bioinformatics
-
Adaptive evolution of molecular structures
Research Summary
Our lab studies the three-dimensional structures of macromolecular complexes by integrating both experimental and bioinformatic methods from the fields of X-ray crystallography, structural bioinformatics, and evolutionary theory. Our previous research has concentrated on the biophysical basis of sequence-specific recognition of unusually structured nucleic acids (such as ssDNAs and ssRNAs) and on the evolution of proteins involved in this important biological function.
Bayesian and likelihood methods for structural comparison and analysis
Superpositioning macromolecular structures is an essential tool in structural bioinformatics and is used routinely in the fields of NMR, X-ray crystallography, protein folding, molecular dynamics, rational drug design, and structural evolution. Superpositioning allows comparison of structures by fitting their atomic coordinates to each other as closely as possible. Interpretation of a superposition relies upon the accuracy of the estimated orientations of the molecules, and thus reliable and robust superpositioning tools are a critical component of structural analysis and comparison.
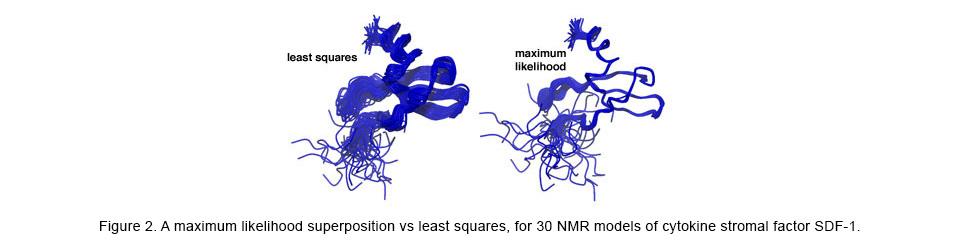
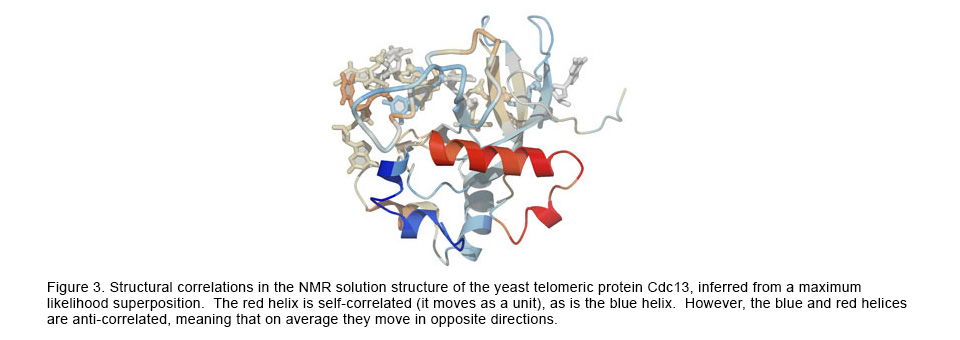
The structural superposition problem has classically been solved with the standard statistical optimization method of least-squares (LS). However, LS can provide misleading and inaccurate results in theory and in practice. To correct for the shortcomings of LS, we have applied likelihood and Bayesian techniques to the superposition problem, resulting in much more accurate superpositions and analyses of the complex correlations among the atoms within macromolecules. Visit this website for more information.
Future Goals and Research
The lab's long-term scientific goals lie in developing precise molecular understandings of the function of macromolecular assemblies, an endeavor which ultimately must be informed by evolutionary knowledge. Currently, the dominant paradigm in structural biology is neutral evolutionary theory, which assumes that the differences among homologous proteins are unimportant for their functions. However, according to the theory of natural selection, differences among proteins can be important for function. Thus, for a full understanding of the relationship between macromolecular function and structure, we consider it essential to explicitly incorporate the modern developments in population genetics regarding natural selection. Conversely, structural knowledge can also inform evolutionary inferences. Implementation of these ideas requires rigorous bioinformatic techniques and modern phylogenetic methods.
 One ongoing research project involves "protein resurrection" methods, in which multiple ancient and extinct proteins are recreated in the lab, assayed experimentally for enzymatic activity, and their atomic resolution structures determined by crystallography. One of the goals of this research is to create a movie in which we can watch how the three-dimensional structure of a macromolecule has evolved in different lineages via point mutations, with each change correlated with changes in the molecule's biochemical function. These "structo-evo" studies will shed light on important structure-function questions, including possibilities for the rational design of proteins with novel functions and for understanding how changes in proteins can affect their function and structures.
One ongoing research project involves "protein resurrection" methods, in which multiple ancient and extinct proteins are recreated in the lab, assayed experimentally for enzymatic activity, and their atomic resolution structures determined by crystallography. One of the goals of this research is to create a movie in which we can watch how the three-dimensional structure of a macromolecule has evolved in different lineages via point mutations, with each change correlated with changes in the molecule's biochemical function. These "structo-evo" studies will shed light on important structure-function questions, including possibilities for the rational design of proteins with novel functions and for understanding how changes in proteins can affect their function and structures.
Recent Publications
-
Trieu MM, Devine EL, Lamarche LB, Ammerman AE, Greco JA, Birge RR, Theobald DL, and Oprian DD. "Purification and Characterization of RhoGC, a Retinylidene/Guanylyl Cyclase Fusion Protein from Blastocladiella emersonii." In press, Journal of Biological Chemistry.
-
Nguyen V, Wilson C, Hoemberger M, Stiller JB, Agafonov RV, Kutter S, English J, Theobald DL, Kern D. (2017) "Evolutionary drivers of thermoadaptation in enzyme catalysis." Science. 2017 Jan 20;355(6322):289-294.
-
Devine EL, Theobald DL, Oprian DD. (2016) "Relocating the Active-Site Lysine in Rhodopsin: 2. Evolutionary Intermediates." Biochemistry. 2016 Aug 30;55(34):4864-70.
-
Steindel PA, Chen EH, Wirth JD and Theobald DL (2016). "Gradual neofunctionalization in the convergent evolution of trichomonad lactate and malate dehydrogenases." Protein Sci. 2016 Feb 17. doi: 10.1002/pro.2904.
-
Theobald DL (2016). "Presenilin adopts the ClC channel fold." Protein Sci. 2016 Mar 12. doi: 10.1002/pro.2919.
-
Wilson C, Agafonov RV, Hoemberger M, Kutter S, Zorba A, Halpin J, Buosi V, Otten R, Waterman D, Theobald DL and Kern D (2015). "Kinase dynamics. Using ancient protein kinases to unravel a modern cancer drug's mechanism." Science 347(6224): 882-886.
-
Hamelryck T, Boomsma W, Ferkinghoff-Borg J, Frellsen J, Haslett J, Kent JT, Mardia KV, and Theobald DL. "Proteins, physics, and probabilities: An outline of a Bayesian formulation of the protein folding problem." Geometry Driven Statistics. Ed. John T. Kent and Ian Dryden. Chichester: John Wiley and Sons, 2015. 356-376.
-
Boucher JI, Jacobowitz JR, Beckett BC, Classen S and Theobald DL (2014). "An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases." Elife. 2014 Jun 25;3. doi: 10.7554/eLife.02304.
-
Mackin KA, Roy RA, and Theobald DL (2014) "An empirical test of convergent evolution in rhodopsins." Mol Biol Evol. 2014 Jan;31(1):85-95.
-
Ni L, Bronk P, Chang EC, Lowell AM, Flam JO, Panzano VC, Theobald DL, Griffith LC and Garrity PA (2013) "A gustatory receptor paralog controls rapid warmth avoidance in Drosophila." Nature 500(7464):580-584. doi:10.1038/nature12390,
-
Devine EL, Oprian DD and Theobald DL (2013) "Relocating the active-site lysine in rhodopsin and implications for evolution of the retinylidene proteins." Proceedings of the National Academy of Sciences USA 110(33):13351-13355.
-
Lyumkis D, Brilot AF, Theobald DL and Grigorieff N (2013) "Likelihood-based classification of cryo-EM images using FREALIGN." Journal of Structural Biology 183(3): 377-388.
-
Mardia KV, Fallaize CJ, Barber S, Jackson RM and Theobald DL (2013) "Bayesian alignment of similarity shapes." The Annals of Applied Statistics 7(2): 989-1009.
-
Theobald DL and Steindel PA (2012) "Optimal simultaneous superpositioning of multiple structures with missing data." Bioinformatics 28 (15): 1972-1979.
-
Theobald DL (2012) "Likelihood and empirical Bayes superpositions of multiple macromolecular structures." In Bayesian Methods in Structural Bioinformatics, Thomas Hamelryck, Kanti V. Mardia, and Jesper Ferkinghoff-Borg, Editors, Statistics for Biology and Health Series, Springer Verlag, New York.
-
Theobald DL (2011) "On universal common ancestry, sequence similarity, and phylogenetic structure: The sins of P-values and the virtues of Bayesian evidence." Biol Direct. 2011 Nov 24;6(1):60.
-
Kene Piasta, Douglas L. Theobald, and Christopher Miller (2011) "Potassium-selective block of barium permeation through single KcsA channels." Journal of General Physiology 138(4): 421-436.
-
Pu Liu, Dimitris K. Agrafiotis, and Douglas L. Theobald (2010) "Fast determination of the optimal rotational matrix for macromolecular superpositions." Journal of Computational Chemistry 31(7): 1561–1563.
-
Theobald DL (2010). "A formal test of the theory of universal common ancestry." Nature 2010 May 13; 465(7295):219-22.
-
Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL and Garrity PA (2010). "Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception." Nature. 2010 Mar 25;464(7288):597-600.
-
Theobald DL (2010). "Likelihood and empirical Bayes superpositions of multiple macromolecular structures." Bayesian methods in structural bioinformatics. Ed. Hamelryck T, Mardia KV, and Ferkinghoff-Borg J. New York: Springer Verlag, 2010.
-
Theobald DL and Miller C (2010). "Membrane transport proteins: Surprises in structural sameness." Nature Structural & Molecular Biology2010 Jan;17(1):2-3.
-
Theobald DL (2009). A nonisotropic Bayesian approach to superpositioning multiple macromolecules.. Proc. of the 28th Leeds Annual Statistical Research (LASR) Workshop, "Statistical Tools for Challenges in Bioinformatics". University of Leeds, UK: 2009.
-
Theobald DL and Wuttke DS (2008). "Accurate structural correlations from maximum likelihood superpositions." PLoS Comput Biol. 2008 Feb;4(2):e43.
-
Theobald DL, Darity WA (2007). "Punctuated equilibrium." International Encyclopedia of the Social Sciences. Second Edition ed. 1 vols. 2007.
-
Theobald DL and Wuttke DS. "Empirical Bayes hierarchical models for regularizing maximum likelihood estimation in the matrix Gaussian Procrustes problem." Proc Natl Acad Sci U S A 103. 49 (2006): 18521-7.
-
Theobald DL and Wuttke DS (2006). "THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures." Bioinformatics 22.17: 2171-2.
-
Theobald DL and Wuttke DS (2005). "Divergent evolution within protein superfolds inferred from profile-based phylogenetics." J Mol Biol 354. 3: 722-37.
-
Theobald DL (2005). "Rapid calculation of RMSDs using a quaternion-based characteristic polynomial." Acta Crystallogr A. 2005 Jul;61(Pt 4):478-80.
-
Mitton-Fry RM, Anderson EM, Theobald DL, Glustrom LW, and Wuttke DS (2004). "Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13." J Mol Biol 338. 2 (2004): 241-55.
-
Theobald DL and Wuttke DS (2004). "Prediction of multiple tandem OB-fold domains in telomere end-binding proteins Pot1 and Cdc13."Structure. 2004 Oct;12(10):1877-9.
-
Theobald DL and Schultz SC (2003). "Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes." Embo J 22. 16 (2003): 4314-24.
-
Theobald DL, Cervantes RB, Lundblad V, and Wuttke DS (2003). "Homology among telomeric end-protection proteins." Structure. 2003 Sep;11(9):1049-50.
-
Theobald DL, Mitton-Fry RM, and Wuttke DS (2003). "Nucleic acid recognition by OB-fold proteins." Annual Review of Biophysics and Biomolecular Structure 32. (2003): 115-33.