2018 Research Highlights

The SciComm Lab Team: Joia, Fellow, Physics PhD candidate; Gabe, Fellow, Physics PhD candidate; Steve, Fellow, Rodal Lab Postdoc; Jennifer, Fellow, Chemistry PhD candidate; Tim, Fellow, Griffith Lab Postdoc; Madelen, Fellow, Neuroscience PhD candidate.
The MRSEC Education, Outreach and Diversity Director (A. Olivier-Mason) developed a new resource called the Science Communication Lab (SciComm Lab). Modeled after a successful Center with the same name at MIT, the Brandeis SciComm Lab provides one-on-one coaching and just-in-time workshops to students and postdocs at Brandeis. The trained graduate student and postdoc communication fellows support the Brandeis scientific community with the skills they need to communicate the rationale, process and results of their incredible science to any audience in written, spoken or visual form.

This year, the MRSEC hosted a large one-day field trip of Waltham High School students as part of an extension of the partnership between Brandeis and Waltham High School. The district has a high percentage of students from backgrounds underrepresented in the sciences, and the MRSEC has several on-going programs with their teachers and students to help broaden participation in STEM. For the field trip, 120 sophomores and juniors enrolled in either sheltered English immersion or C2 (low-level) biology and chemistry classes came to Brandeis. Many of the students were new to the country, and/or for whom English is a second language, and/or are not typically aware of educational opportunities beyond high school. The goal was to introduce Brandeis and ignite their interests in science. More than 25 Brandeis scientists fluent in Spanish, French and Portuguese volunteered to lead small-group science discussions in those languages. The powerful event included talks delivered by scientists from URM and immigrant backgrounds.
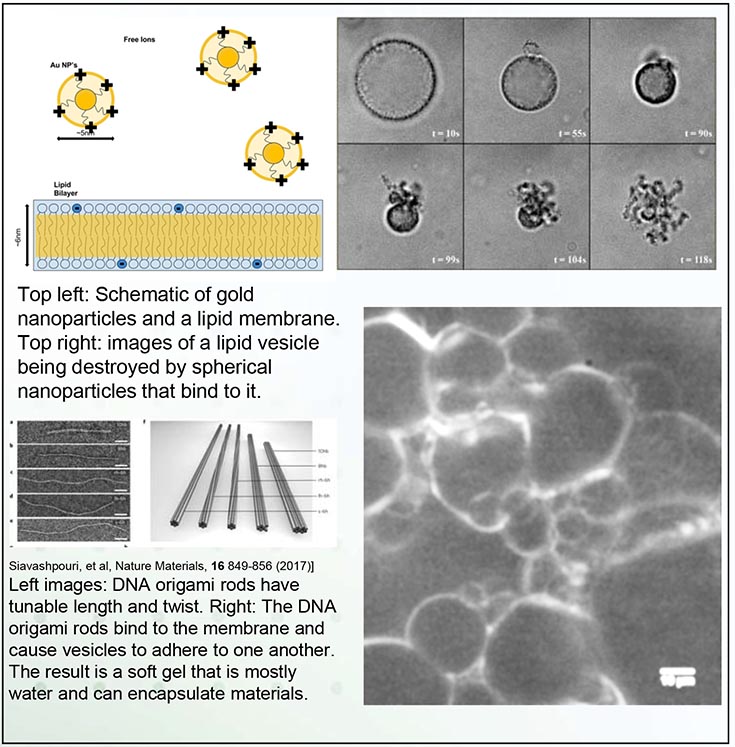
Top left: Schematic of gold nanoparticles and a lipid membrane. Top right: images of a lipid vesicle being destroyed by spherical nanoparticles that bind to it. Left images: DNA origami rods have tunable length and twist. Right: The DNA origami rods bind to the membrane and cause vesicles to adhere to one another. The result is a soft gel that is mostly water and can encapsulate materials.
In a cell membrane, lipids and proteins work in tandem to provide shape, structure and protection for the cell. These biological examples inspire applications in which membranes could form artificial adaptive materials that encapsulate and deliver materials. Achieving this goal requires that we first understand the basic principles for remodeling membranes.
In this MRSEC study, we made simplified cell membranes (called vesicles). We exposed them to nanoparticles that adhered to the membranes with a controllable binding energy. When the binding energy was strong enough to overcome the membrane-bending energy, the membranes suffered a surprising and dramatic inversion of the shape, leaving an octopus-like structure. Computer simulations helped to identify criteria for this process to occur. Using DNA origami, we also made particles with different shapes (here, twisted rods) that bound to membranes and yielded different structures. This approach will help us predict how protein/particle shape drive membrane remodeling.
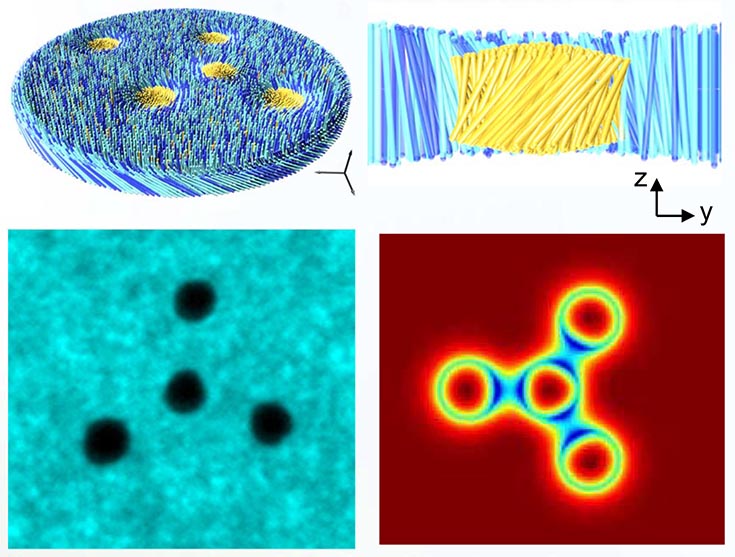
Top: Schematics of membrane rafts from above and from the side. Bottom Left: Fluorescence image of four rafts assembled in a tetramer formation. Bottom Right: Heatmap showing a numerical calculation for the degree of tilt for a similar configuration.
Particles suspended in liquid membranes interact based on their deformation of the background membrane rather than their inherent properties. This leads to many complex interactions which have not been fully explored. Recent experiments with self-assembled rafts in colloidal membranes have found that rafts twist the membrane around them and can switch between a right-handed or left-handed twist configuration. Rafts in like states have repulsive interactions, while unlike states lead to metastably bound pairs. Theory has shown that both states are metastable and confirmed that these interactions are driven by minimizing membrane deformation.
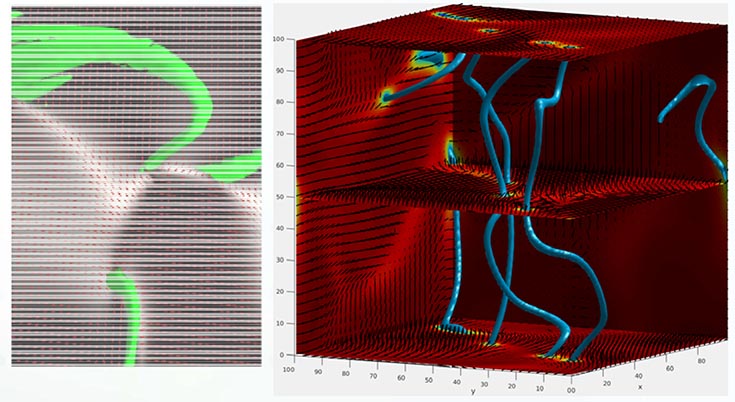
Structure of a 3D active nematic. (Left) Light sheet microscopy image, with disclination lines shown by green tubes. (Right) Simulation, with disclination lines shown by blue tubes. The director field (lines) and nematic order parameter (colors) are shown in 3 planes.
Active materials are continuously driven out of equilibrium by energy input at the particle scale, potentially enabling applications that are not possible with equilibrium materials. However, most current active materials are limited to 2D, whereas practical applications require 3D systems. The MRSEC has developed the first truly 3D active nematic by combining microtubules, molecular motors and passive rod-like viruses. Using light sheet microscopy we are able to visualize the system at unprecedented resolution (a snapshot is shown on the right), showing that the system is spanned by a complex dynamical network of disclination lines (localized regions of disorder). Theoretical calculations (bottom image on the right) are being used to identify the mechanisms controlling the formation and dynamics of disclination lines.
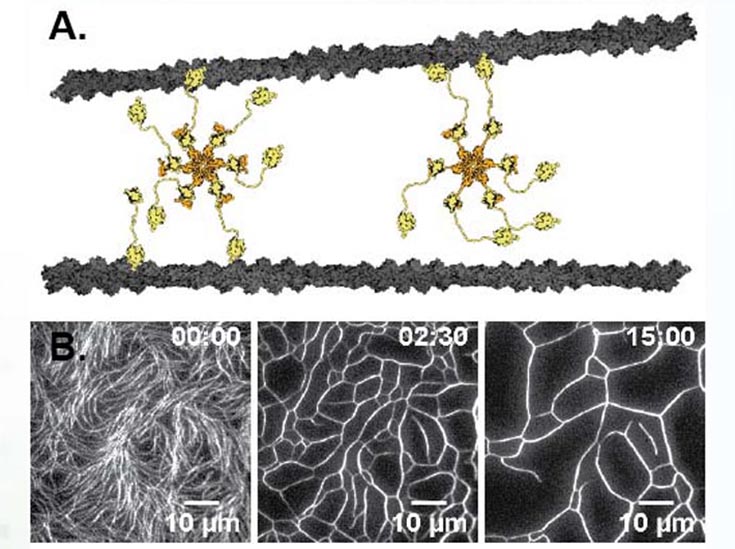
(A) The hexameric structure of the Srv2/Abp1 actin crosslinker allows for non‐canonical interactions. (B) Actin filaments are confined to a 2d interface re‐organized by Abp1/Srv2 into a coarsened cellular network.
One of the goals of the MRSEC is to develop new active materials built from biomaterial components. The investigators have now made a truly macroscopic (millimeter sized) 3D contractile active gel composed of actin crosslinked with the hexameric Srv2/Abp1 complex, that coarsens an entangled network of actin into a cellular foam like network. This actin scaffold is being combined with PRC-crosslinked extensile microtubule bundles to make a truly multicomponent composite active material with both contractile and extensile activity. Theoretical analysis in ongoing to understand the coarsening dynamics of the actin scaffold in the presence of the Srv2/Abp1 complex.
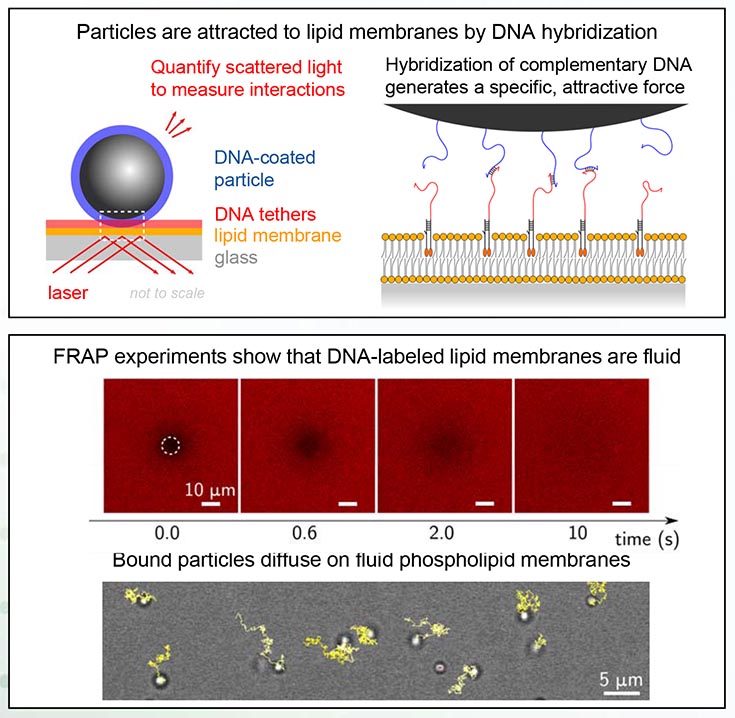
Top image, titled: Particles are attracted to lipid membranes by DNA hybridization." Lower image is titled "FRAP experiments show that DNA-labeled lipid membranes are fluid." Bound particles diffuse on fluid phospholipid membranes. Top image, titled: Particles are attracted to lipid membranes by DNA hybridization, has 2 diagrams. Left is labeled "Quantify scattered light to measure interactions." Parts of the diagram are labeled as: DNA-coated particle, DNA tethers, lipid, membrane glass and laser. Right diagram is titled "Hybridization of complementary DNA generates a specific attractive force." Lower image is titled "FRAP experiments show that DNA-labeled lipid membranes are fluid." Text below says: "Bound particles diffuse on fluid phospholipid membranes."
Using DNA nanotechnology, we investigate how micrometer-scale particles spontaneously organize on lipid membranes to fold them into remarkable nanostructures. We have successfully functionalized particles and lipid membranes with DNA tethers, and built a laser-based microscope to measure the particle-membrane interactions with femtonewton resolution.
In future MRSEC IRG 1 experiments, we will characterize particle-membrane interactions and assess the effects of membrane fluidity and ligand-receptor affinity on spontaneous organization of membrane-bound particles. This work may enable new self-assembling, membranebased materials with remarkable optical properties, such as structural coloration.
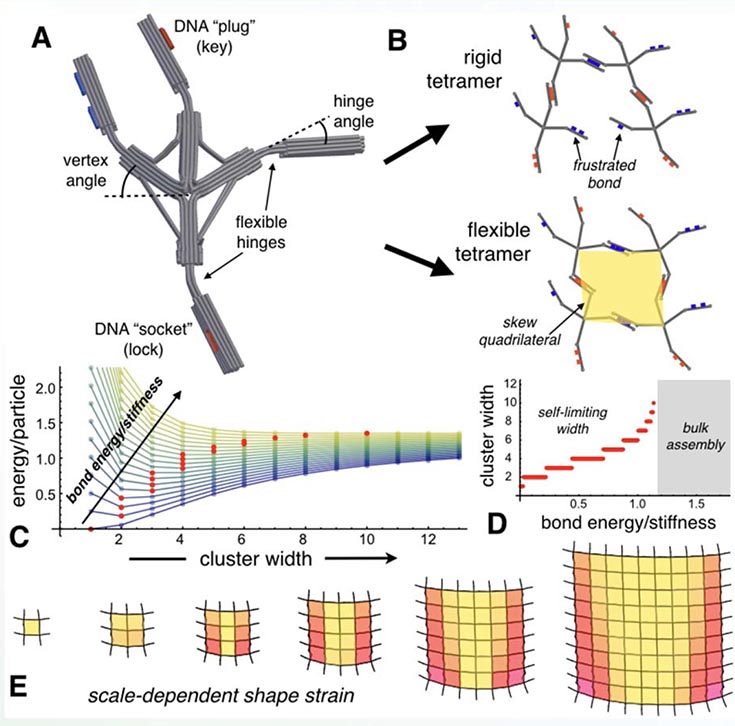
(A) Finite-element simulation of DNA origami vertex particle, cylinders depicting base-paired double strands; (B) Angular geometry frustrates bonds and requires cluster deformation; Size-dependent energy/particle (C) predicts self-limiting domains (D) due to the accumulation of shape strain (E).
The assembly of identical sub-units into larger, yet finitesized, superstructures is ubiquitous and functionally vital in biology, from photonic nanostructures in butterfly wings to size-regulated protein fibers. An emerging paradigm to achieve finite architectures in synthetic systems is the incorporation of geometric frustration into the assembly process. Like an ill-fitting jigsaw puzzle, such assemblies require mechanical deformation of building blocks, which in turn, feeds back into self-limiting equilibrium domains.
To realize particles with the control over inter-particle misfit and shape deformability needed for size-selective assembly, a new class of ‘frustrated vertex’ particles was designed for synthesis via state-of-the-art DNA origami techniques. Angular geometry of vertices and pre-bent hinges frustrate the ‘lock and key’ bonds between particle arms, leading to skewed local packing incompatible uniform planar tiling. Computational modeling verifies the accumulation of hinge bending with cluster size, which gives rise to equilibrium domains whose finite width is tunable via strength of DNA-mediated bonds.

