Suzanne Paradis
 Professor of Biology
Professor of Biology
Research Description
Molecular Mechanisms of Synapse Development
The complex circuitry of the mammalian brain enables the execution of fundamental cognitive processes such as learning, speech, and memory. Neural circuits are assembled via specialized sites of cell-cell contact and communication between neurons termed synapses. Aberrant synapse development can have pathological consequences for circuit function as demonstrated by the manifestation of devastating neurological impairments, including epilepsy and autism spectrum disorders. The aim of our research is to define the molecular program that underlies both excitatory and inhibitory synapse development with the goal of contributing to a greater understanding of neural circuit formation and function.
While biochemical and candidate gene approaches have led to the identification of a large number of molecules that function at the synapse, the process of synapse development itself remains poorly understood. Some of the critical questions in the field include:
- Which proteins are required for excitatory and inhibitory synapse development and what is their mechanism of action?
- At which specific step in synapse development is the activity of each protein required?
- How does a neuron maintain the correct balance of excitatory and inhibitory synapses in order to function appropriately within a neural circuit?
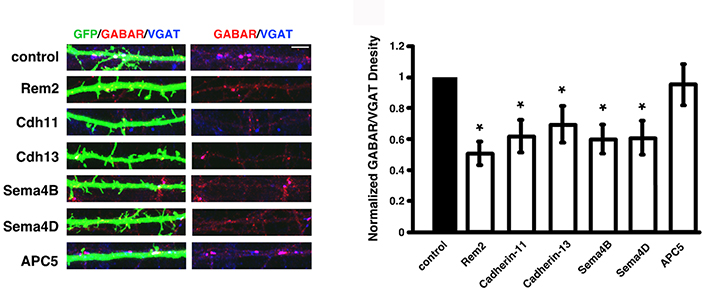
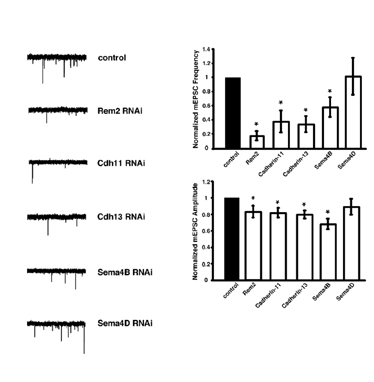 We have begun to address these important questions using a novel, forward genetic RNA interference (RNAi)-based screen in cultured hippocampal neurons (Fig. 1) that has identified new molecules required for synapse development. Thus far, we have isolated five new genes that are required for the proper development of excitatory and/or inhibitory synapses (Figs. 2&3).
We have begun to address these important questions using a novel, forward genetic RNA interference (RNAi)-based screen in cultured hippocampal neurons (Fig. 1) that has identified new molecules required for synapse development. Thus far, we have isolated five new genes that are required for the proper development of excitatory and/or inhibitory synapses (Figs. 2&3).
To investigate the function of the genes isolated in this and future screens, we utilize a combination of molecular, biochemical, and electrophysiological approaches in primary cultures of hippocampal neurons, organotypic hippocampal slice, acute hippocampal slice, and mouse models. In addition, as only 30% of genes in the mammalian genome have an ascribed function, a complete understanding of synapse development and circuit function depends on identifying the full complement of molecules that mediate these important processes. Thus, future screens will focus on isolating molecules that function to regulate the development of either excitatory or inhibitory synapses, or that act as general promoters of synaptic development.
Selected Publications
- Experience-Dependent Development of Dendritic Arbors in Mouse Visual Cortex. Richards SEV, Moore AR, Nam AY, Saxena S, Paradis S, Van Hooser SD. J Neurosci. 2020 Aug 19;40(34):6536-6556. doi: 10.1523/JNEUROSCI.2910-19.
2020. - TDP-43 dysfunction restricts dendritic complexity by inhibiting CREB activation and altering gene expression. Herzog JJ, Xu W, Deshpande M, Rahman R, Suib H, Rodal AA, Rosbash M, Paradis S. Proc Natl Acad Sci U S A. 2020 May 26;117(21):11760-11769. doi: 10.1073/pnas.1917038117.
-
The Metastasis Suppressor Protein Nme1 Is a Concentration-Dependent Modulator of Ca2+/Calmodulin-Dependent Protein Kinase II. Royer L, Shangraw K, Herzog JJ, Pouvreau S, Marr MT 2nd, Paradis S. Biochemistry. 2019 Jun 18;58(24):2710-2714.
-
Stable memory and computation in randomly rewiring neural networks. Acker D, Paradis S, Miller P. J Neurophysiol. 2019 Jul 1;122(1):66-80.
- Steinmetz, C. C., V. Tatavarty, K. Sugino, Y. Shima, A. Joseph, H. Lin, M. Rutlin, M. Lambo, C. M. Hempel, B. W. Okaty, S. Paradis, S. B. Nelson and G. G. Turrigiano (2016). "Upregulation of mu3A Drives Homeostatic Plasticity by Rerouting AMPAR into the Recycling Endosomal Pathway." Cell Rep 16(10): 2711-2722.
- Ghiretti AE, Moore AR, Brenner RG, Chen LF, West AE, Lau NC, Van Hooser SD, Paradis S (2014). "Rem2 is an activity-dependent negative regulator of dendritic complexity in vivo." J Neurosci 34(2): 392-407.
- Ghiretti AE and Paradis S (2014). "Molecular mechanisms of activity-dependent changes in dendritic morphology: role of RGK proteins." Trends Neurosci 37(7): 399-407.
- Raissi AJ, Scangarello FA, Hulce KR, Pontrello JK, Paradis S (2014). "Enhanced potency of the metalloprotease inhibitor TAPI-2 by multivalent display." Bioorg Med Chem Lett 24(8): 2002-2007.
- Ghiretti AE, Kenny K, Marr MT 2nd, Paradis S. (2013). "CaMKII-dependent phosphorylation of the GTPase Rem2 is required to restrict dendritic complexity." J Neurosci 33(15): 6504-6515.
- Kuzirian MS, Moore AR, Staudenmaier EK, Friedel RH, Paradis S (2013). "The class 4 semaphorin Sema4D promotes the rapid assembly of GABAergic synapses in rodent hippocampus." J Neurosci 33(21): 8961-8973.
- Moore AR, Ghiretti AE, Paradis S (2013). "A loss-of-function analysis reveals that endogenous Rem2 promotes functional glutamatergic synapse formation and restricts dendritic complexity." PLoS One 8(8): e74751.
- Raissi AJ, Staudenmaier EK, David S, Hu L, Paradis S (2013). "Sema4D localizes to synapses and regulates GABAergic synapse development as a membrane-bound molecule in the mammalian hippocampus." Mol Cell Neurosci 57: 23-32.
- Zeng M, Kuzirian MS, Harper L, Paradis S, Nakayama T, Lau NC (2013). "Organic small hairpin RNAs (OshR): a do-it-yourself platform for transgene-based gene silencing." Methods 63(2): 101-109.
- Kuzirian MS, Paradis S. (2011) "Emerging themes in GABAergic synapse development." Prog Neurobiol. 2011 Jul 20;95(1):68-87.
- Ghiretti AE, Paradis S (2011). "The GTPase Rem2 regulates synapse development and dendritic morphology." Dev Neurobiol 2011 May;71(5):374-89.
- Paradis S*, Harrar DB*, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME (2007) "An RNAi-based Approach Identifies New Molecules Required for Glutamatergic and GABAergic Synapse Development." Neuron 53: 217-232.
- An immunocytochemistry - based asay of inhibitory synapse density demonstrates that Rem2, Cadherin-11, Cadherin-13, Sema4B or Sema4D are required for the proper development of inhibitory synapses.
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME (2006). "Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number." Science 311: 1008-1012.
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME (2005). "The Rac1-GEF Tiam1 Couples the NMDA Receptor to the Activity-Dependent Development of Dendritic Arbors and Spines." Neuron 45: 525-538.
- Paradis S, Sweeney ST, Davis GW (2001). "Homeostatic Control of Presynaptic Release is Triggered by Postsynaptic Membrane Depolarization." Neuron 30: 737-749.
- Davis GW, Eaton B, Paradis S (2001). "Synapse Formation Revisited." Nat. Neurosci. 4: 558-560.
*authors contributed equally